Date: Tue, 15 Dec 2020 13:03:19 +0530
Can you confirm if you are using radiopt=0 (default is =1) for PB
calculation?
Also, try to run mmpbsa.py in serial mode to check if it runs without
errors/.
Best Regards
Elvis
On Tue, 15 Dec 2020 at 12:56, Vaibhav Dixit <vaibhavadixit.gmail.com> wrote:
> Dear All and Elvis,
> The GB calculation did work after recompling with the suggested changes.
> But the PB calculation is failing with the following error messages.
> Inspection of the output file shows that it did not find radii for the S
> atom which has type "Y1" as defined by the MCPB.py tool which I had used
> for building parameters for the co-factor.
> Thus I'm wondering does this require adding similar lines in the PB
> section of the mdread2.F90 file?
> Please help me understand how should I try to fix this error?
> Thank you and best regards.
>
> tail of the _MMPBSA_complex_pb.mdout.0 file
> Frozen or restrained atoms:
> ibelly = 0, ntr = 0
>
> Energy minimization:
> maxcyc = 1, ncyc = 10, ntmin = 1
> dx0 = 0.01000, drms = 0.00010
> PB Bomb in pb_aaradi(): No radius assigned for atom 5179 SG Y1
>
> (base) [exx.c107739 mmpbsa-6701-tyr]$ MMPBSA.py -O -i mmpbsa-decomp.in -o
> mmpbs-rouf-6701-tyr.dat -do decomp-rouf-6701-tyr.dat -sp
> ../rouf-6701-tyr-solv.prmtop -cp ../rouf-6701-tyr-dry.prmtop -lp
> ../tyr-dry.prmtop -y ../rouf-6701-tyr-solv-prod300.nc -rp
> ../rouf-6701-nolig-dry.prmtop
> Loading and checking parameter files for compatibility...
> /home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py:603:
> UserWarning: receptor_mask overwritten with default
>
> warnings.warn('receptor_mask overwritten with default\n')
> cpptraj found! Using /home/exx/Downloads/amber20/bin/cpptraj
> sander found! Using /home/exx/Downloads/amber20/bin/sander
> Preparing trajectories for simulation...
> 20 frames were processed by cpptraj for use in calculation.
>
> Running calculations on normal system...
>
> Beginning GB calculations with /home/exx/Downloads/amber20/bin/sander
> calculating complex contribution...
> calculating receptor contribution...
> calculating ligand contribution...
>
> Beginning PB calculations with /home/exx/Downloads/amber20/bin/sander
> calculating complex contribution...
> File "/home/exx/Downloads/amber20/bin/MMPBSA.py", line 100, in <module>
> app.run_mmpbsa()
> File
> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py",
> line 218, in run_mmpbsa
> self.calc_list.run(rank, self.stdout)
> File
> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
> line 82, in run
> calc.run(rank, stdout=stdout, stderr=stderr)
> File
> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
> line 428, in run
> error_list = [s.strip() for s in out.split('\n')
> TypeError: a bytes-like object is required, not 'str'
> Exiting. All files have been retained.
>
>
>
> On Tue, Dec 15, 2020 at 2:39 AM Elvis Martis <elvis_bcp.elvismartis.in>
> wrote:
>
>> Hi Vaibhav
>> I could reproduce the error.
>> The reason for it is, I edited the mdread2.F90 file using notepad++.
>> Apparently, the UTC-8 Unicode in windows has some issues and seems
>> incompatible with Linux, and therefore, the unknown characters.
>>
>> I would recommend, you do a fresh install, untar the AmberTools20.bz2
>> file and go to AmberTools/src/sander and add (i suggest typing it) the
>> required lines in the mdread2.F90 using any editor in Linux itself.
>> It should solve the problem, as I could compile it.
>> Best Regards
>> Elvis
>>
>>
>>
>> On Mon, 14 Dec 2020 at 19:55, Elvis Martis <elvis_bcp.elvismartis.in>
>> wrote:
>>
>>> I will try compile amber20 Using this very mdread2.F90 file and try to
>>> reproduce this error.
>>> I will get back to you asap
>>>
>>> On Monday, December 14, 2020, Vaibhav Dixit <vaibhavadixit.gmail.com>
>>> wrote:
>>>
>>>> Dear Elivs,
>>>> This file is giving me the same error.
>>>> I'm not sure if it is related to the file getting transferred over the
>>>> internet which might lead to strange characters and sort of things.
>>>> Please do let me know if you find a way to fix that.
>>>> thanks
>>>>
>>>> (base) [exx.c107739 build]$ make install
>>>> [ 1%] Built target ucpp
>>>> [ 1%] Built target dft_scalar
>>>> [ 1%] Built target fftw3_threads_obj
>>>> [ 1%] Built target reodft
>>>> [ 1%] Built target simd_support
>>>> [ 1%] Built target rdft_scalar_r2r
>>>> [ 1%] Built target rdft_sse2_codelets
>>>> [ 2%] Built target fftw_kernel
>>>> [ 2%] Built target rdft_scalar
>>>> [ 5%] Built target rdft_scalar_r2cb
>>>> [ 7%] Built target rdft_scalar_r2cf
>>>> [ 8%] Built target dft_scalar_codelets
>>>> [ 12%] Built target dft_sse2_codelets
>>>> [ 13%] Built target rdft
>>>> [ 15%] Built target fftw_api
>>>> [ 16%] Built target dft
>>>> [ 16%] Built target fftw
>>>> [ 17%] Built target libfftw_bench
>>>> [ 17%] Built target fftw_benchmark_common_obj
>>>> [ 17%] Built target fftw_benchmark
>>>> [ 17%] Built target fftw_wisdom
>>>> [ 18%] Built target dispatch
>>>> [ 18%] Built target netcdf3
>>>> [ 18%] Built target netcdf
>>>> [ 18%] Built target ncgen
>>>> [ 18%] Built target ncgen3
>>>> [ 18%] Built target nccopy
>>>> [ 19%] Built target ncdump
>>>> [ 20%] Built target netcdff
>>>> [ 20%] Built target xblas_build
>>>> [ 22%] Built target blas
>>>> [ 28%] Built target lapack
>>>> [ 29%] Built target arpack
>>>> [ 29%] Built target boost_build
>>>> [ 29%] Built target amber_common
>>>> [ 30%] Built target libpbsa
>>>> [ 30%] Built target sff_lex
>>>> [ 30%] Built target dsarpack_obj
>>>> [ 32%] Built target rism
>>>> [ 32%] Built target sff_rism_interface
>>>> [ 32%] Built target sff
>>>> [ 33%] Built target gbnsr6
>>>> [ 33%] Built target cifparse
>>>> [ 34%] Built target addles
>>>> [ 36%] Built target sqm_common
>>>> [ 36%] Built target libsqm
>>>> [ 37%] Built target sff_fortran
>>>> [ 37%] Built target sander_rism_interface
>>>> Scanning dependencies of target sander_base_obj
>>>> make[2]: Warning: File `../AmberTools/src/sander/mdread2.F90' has
>>>> modification time 12412 s in the future
>>>> [ 37%] Building Fortran object
>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1567.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> else if (atomicnumber .eq. 26) then
>>>> 1
>>>> Warning: Nonconforming tab character at (1)
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1568.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 !
>>>> \xC2\xA0Fe ra
>>>> 1
>>>> Error: Invalid character in name at (1)
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1569.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>> x(L165-1+i) = 1
>>>> 1
>>>> Error: Invalid character in name at (1)
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1630.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> else if (atomicnumber .eq. 26) then
>>>> 1
>>>> Warning: Nonconforming tab character at (1)
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1631.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 ! \xC2\xA0Fe
>>>> radius = 1.3
>>>> 1
>>>> Error: Invalid character in name at (1)
>>>>
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1632.1:
>>>> Included at
>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>
>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 x(L165-1+i) =
>>>> 1.39d0 + 1.
>>>> 1
>>>> Error: Invalid character in name at (1)
>>>> make[2]: ***
>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o] Error 1
>>>> make[1]: *** [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all]
>>>> Error 2
>>>> make: *** [all] Error 2
>>>> (base) [exx.c107739 build]$
>>>>
>>>> On Mon, Dec 14, 2020 at 7:30 PM Elvis Martis <elvis_bcp.elvismartis.in>
>>>> wrote:
>>>>
>>>>> Hi Vaibhav
>>>>> The problem was that tabs and indentations were not correct.
>>>>> Also, the x(L165-1+i) = 1.386d0 + 1.4d0 was changed to x(L165-1+i)
>>>>> = 1.39d0 + 1.4d0
>>>>> This time I made sure they are in the right place.
>>>>> I hope this time it will OK
>>>>> Best Regards
>>>>> Elvis
>>>>>
>>>>>
>>>>>
>>>>> On Mon, 14 Dec 2020 at 19:13, Vaibhav Dixit <vaibhavadixit.gmail.com>
>>>>> wrote:
>>>>>
>>>>>> Dear Elvis,
>>>>>> I have copied your file in the sander folder and reran the run_cmake
>>>>>> and make install commands.
>>>>>> But I'm getting new errors after a few steps shown below.
>>>>>> I think it still doesn't like the modifications and syntax like
>>>>>> errors are probably showing up.
>>>>>> Please let me know what modifications should I make now.
>>>>>> thanks for your continued support and suggestions.
>>>>>> Best regards.
>>>>>>
>>>>>> Scanning dependencies of target sander_base_obj
>>>>>> [ 37%] Building Fortran object
>>>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1567.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> else if (atomicnumber .eq. 26) then
>>>>>> 1
>>>>>> Warning: Nonconforming tab character at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1568.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0!
>>>>>> \xC2\xA0Fe rad
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1569.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>> x(L165-1+i) = 1
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1570.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 else
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1571.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>> \xC2\xA0write( 0
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1572.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>> \xC2\xA0call mex
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1630.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> else if (atomicnumber .eq. 26) then
>>>>>> 1
>>>>>> Warning: Nonconforming tab character at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1631.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0!
>>>>>> \xC2\xA0Fe rad
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>>
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1632.1:
>>>>>> Included at
>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>
>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>> x(L165-1+i) = 1
>>>>>> 1
>>>>>> Error: Invalid character in name at (1)
>>>>>> make[2]: ***
>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o] Error 1
>>>>>> make[1]: ***
>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all] Error 2
>>>>>> make: *** [all] Error 2
>>>>>>
>>>>>> On Mon, Dec 14, 2020 at 10:31 PM Elvis Martis <
>>>>>> elvis_bcp.elvismartis.in> wrote:
>>>>>>
>>>>>>> Hi Vaibhav
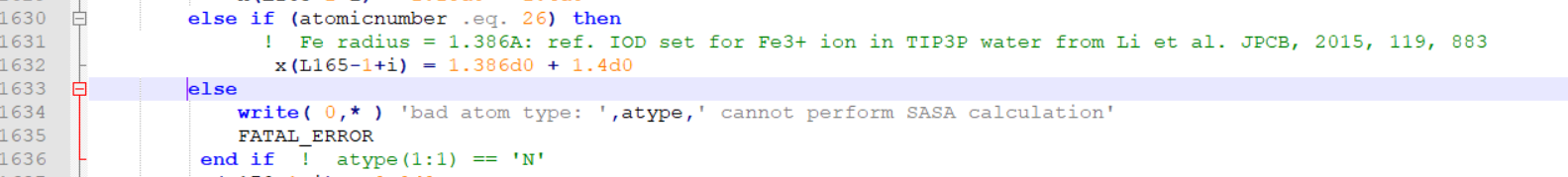
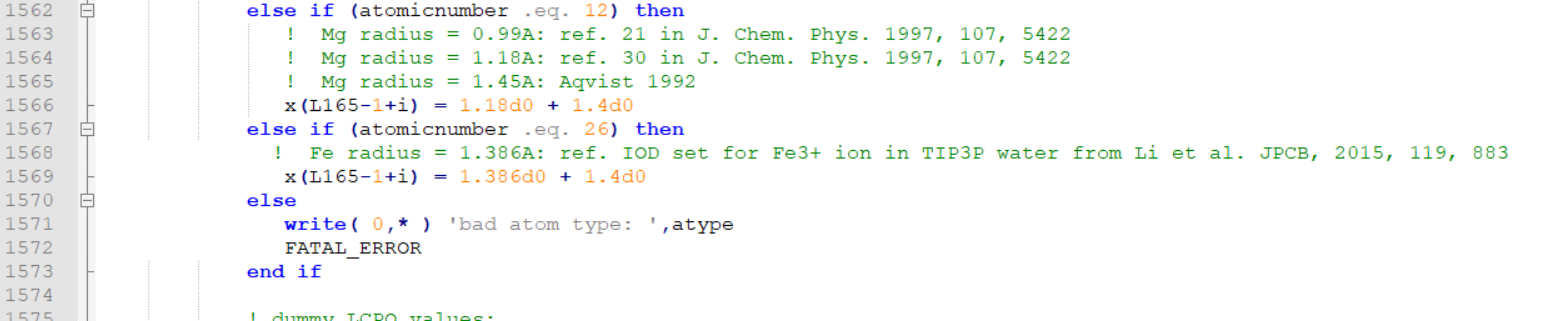
>>>>>>> I guess the error was because of the commenting the "else" line.
>>>>>>> I have edited mdread2.F90 and attached here.
>>>>>>> I am pasting the snapshots of the changes I did at two places.
>>>>>>> [image: image.png]
>>>>>>>
>>>>>>> [image: image.png]
>>>>>>>
>>>>>>> You can directly paste this mread2.F90 in the src/sander and
>>>>>>> recompile.
>>>>>>> Best Regards
>>>>>>> Elvis
>>>>>>>
>>>>>>>
>>>>>>>
>>>>>>> On Mon, 14 Dec 2020 at 18:32, Vaibhav Dixit <vaibhavadixit.gmail.com>
>>>>>>> wrote:
>>>>>>>
>>>>>>>> Dear Elvis,
>>>>>>>> I searched for the igb == 2 and then "bad atom type".
>>>>>>>> I commented out the following lines (as shown).
>>>>>>>> these are line number 1567, 1568 and 1569 in the attached file.
>>>>>>>>
>>>>>>>> ! else
>>>>>>>> ! write( 0,* ) 'bad atom type: ',atype
>>>>>>>> ! FATAL_ERROR
>>>>>>>>
>>>>>>>> Then added the following lines
>>>>>>>> else if (atomicnumber .eq. 23) then
>>>>>>>> ! Fe radius = 1.386A: ref. IOD set for Fe3+ ion in
>>>>>>>> TIP3P
>>>>>>>> water from Li et al. JPCB, 2015, 119, 883
>>>>>>>> x(L165-1+i) = 1.386d0 + 1.4d0
>>>>>>>>
>>>>>>>> These line numbers are 1570, 1571 and 1572 in the attached file.
>>>>>>>>
>>>>>>>> Please let me know if I need to make additional changes in that
>>>>>>>> file.
>>>>>>>> Thank you and best regards.
>>>>>>>>
>>>>>>>>
>>>>>>>> On Mon, Dec 14, 2020 at 9:53 PM Elvis Martis <
>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>> wrote:
>>>>>>>>
>>>>>>>> > Can you point out what changes you did exactly?
>>>>>>>> >
>>>>>>>> >
>>>>>>>> > Best Regards
>>>>>>>> > Elvis
>>>>>>>> >
>>>>>>>> >
>>>>>>>> >
>>>>>>>> > On Mon, 14 Dec 2020 at 18:14, Vaibhav Dixit <
>>>>>>>> vaibhavadixit.gmail.com>
>>>>>>>> > wrote:
>>>>>>>> >
>>>>>>>> > > Dear All and Elivs,
>>>>>>>> > > OK, now I got the file in amber20_src/AmberTools/src/sander.
>>>>>>>> > > I have made the following changes (in bold), and then set DMPI
>>>>>>>> and DCUDA
>>>>>>>> > > both =FALSE and then ran run_make script.
>>>>>>>> > > But I get the error shown at the end.
>>>>>>>> > > Can you please check and help me understand what corrections to
>>>>>>>> me to
>>>>>>>> > these
>>>>>>>> > > lines?
>>>>>>>> > >
>>>>>>>> > > Thanks a lot and best regards.
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> > > x(L165-1+i) = 1.10d0 + 1.4d0
>>>>>>>> > > else if (atomicnumber .eq. 12) then
>>>>>>>> > > ! Mg radius = 0.99A: ref. 21 in J. Chem. Phys.
>>>>>>>> 1997, 107,
>>>>>>>> > > 5422
>>>>>>>> > > ! Mg radius = 1.18A: ref. 30 in J. Chem. Phys.
>>>>>>>> 1997, 107,
>>>>>>>> > > 5422
>>>>>>>> > > ! Mg radius = 1.45A: Aqvist 1992
>>>>>>>> > > x(L165-1+i) = 1.18d0 + 1.4d0
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> > > *# else# write( 0,* ) 'bad atom type:
>>>>>>>> ',atype#
>>>>>>>> > > FATAL_ERROR*
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> > > * else if (atomicnumber .eq. 23) then
>>>>>>>> ! Fe
>>>>>>>> > radius
>>>>>>>> > > = 1.386A: ref. IOD set for Fe3+ ion in TIP3P water from Li et
>>>>>>>> al. JPCB,
>>>>>>>> > > 2015, 119, 883 x(L165-1+i) = 1.386d0 + 1.4d0*
>>>>>>>> > > end if
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> > > *Error with run_cmake script*
>>>>>>>> > > (base) [exx.c107739 build]$ gedit run_cmake &
>>>>>>>> > > [2] 16544
>>>>>>>> > > (base) [exx.c107739 build]$ make install
>>>>>>>> > > [ 0%] Built target ucpp
>>>>>>>> > > [ 0%] Built target fftw3_threads_obj
>>>>>>>> > > [ 1%] Built target fftw_api
>>>>>>>> > > [ 1%] Built target dft
>>>>>>>> > > [ 1%] Built target dft_scalar
>>>>>>>> > > [ 2%] Built target dft_scalar_codelets
>>>>>>>> > > [ 5%] Built target dft_sse2_codelets
>>>>>>>> > > [ 5%] Built target rdft
>>>>>>>> > > [ 5%] Built target rdft_scalar
>>>>>>>> > > [ 7%] Built target rdft_scalar_r2cb
>>>>>>>> > > [ 8%] Built target rdft_scalar_r2cf
>>>>>>>> > > [ 8%] Built target rdft_scalar_r2r
>>>>>>>> > > [ 8%] Built target rdft_sse2_codelets
>>>>>>>> > > [ 9%] Built target fftw_kernel
>>>>>>>> > > [ 9%] Built target reodft
>>>>>>>> > > [ 9%] Built target simd_support
>>>>>>>> > > [ 9%] Built target fftw
>>>>>>>> > > [ 9%] Built target libfftw_bench
>>>>>>>> > > [ 9%] Built target fftw_benchmark_common_obj
>>>>>>>> > > [ 9%] Built target fftw_benchmark
>>>>>>>> > > [ 9%] Built target fftw_wisdom
>>>>>>>> > > [ 9%] Built target fftw_mpi
>>>>>>>> > > [ 9%] Built target fftw_mpi_benchmark
>>>>>>>> > > [ 9%] Built target dispatch
>>>>>>>> > > [ 9%] Built target netcdf3
>>>>>>>> > > [ 9%] Built target netcdf
>>>>>>>> > > [ 10%] Built target ncgen
>>>>>>>> > > [ 10%] Built target ncgen3
>>>>>>>> > > [ 10%] Built target nccopy
>>>>>>>> > > [ 10%] Built target ncdump
>>>>>>>> > > [ 11%] Built target netcdff
>>>>>>>> > > [ 11%] Built target xblas_build
>>>>>>>> > > [ 12%] Built target blas
>>>>>>>> > > [ 15%] Built target lapack
>>>>>>>> > > [ 16%] Built target arpack
>>>>>>>> > > [ 19%] Built target pnetcdf_fortran_obj
>>>>>>>> > > [ 19%] Built target pnetcdf_c_obj
>>>>>>>> > > [ 19%] Built target pnetcdf
>>>>>>>> > > [ 19%] Built target ncvalid
>>>>>>>> > > [ 19%] Built target boost_build
>>>>>>>> > > [ 19%] Built target amber_common
>>>>>>>> > > [ 20%] Built target rism
>>>>>>>> > > [ 20%] Built target libpbsa
>>>>>>>> > > [ 20%] Built target sff_rism_interface
>>>>>>>> > > [ 20%] Built target sff_lex
>>>>>>>> > > [ 20%] Built target dsarpack_obj
>>>>>>>> > > [ 20%] Built target sff
>>>>>>>> > > [ 21%] Built target gbnsr6
>>>>>>>> > > [ 21%] Built target cifparse
>>>>>>>> > > [ 22%] Built target addles
>>>>>>>> > > [ 23%] Built target rism_mpi
>>>>>>>> > > [ 23%] Built target sander_rism_interface
>>>>>>>> > > [ 25%] Built target sqm_common
>>>>>>>> > > [ 25%] Built target libsqm
>>>>>>>> > > [ 26%] Built target sff_fortran
>>>>>>>> > > [ 26%] Building Fortran object
>>>>>>>> > >
>>>>>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>>>>> > >
>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1571.1:
>>>>>>>> > > Included at
>>>>>>>> > >
>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>> > >
>>>>>>>> > > \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0!
>>>>>>>> > \xC2\xA0Fe
>>>>>>>> > > rad
>>>>>>>> > > 1
>>>>>>>> > > Error: Invalid character in name at (1)
>>>>>>>> > >
>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1572.1:
>>>>>>>> > > Included at
>>>>>>>> > >
>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>> > >
>>>>>>>> > > \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>>> > x(L165-1+i)
>>>>>>>> > > = 1
>>>>>>>> > > 1
>>>>>>>> > > Error: Invalid character in name at (1)
>>>>>>>> > > make[2]: ***
>>>>>>>> > >
>>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o]
>>>>>>>> > Error 1
>>>>>>>> > > make[1]: ***
>>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all]
>>>>>>>> > > Error 2
>>>>>>>> > > make: *** [all] Error 2
>>>>>>>> > > [2]+ Done gedit run_cmake
>>>>>>>> > >
>>>>>>>> > > On Mon, Dec 14, 2020 at 8:36 PM Elvis Martis <
>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>> > > wrote:
>>>>>>>> > >
>>>>>>>> > > > One more thing I missed,
>>>>>>>> > > > After you have successfully located the file and edited it
>>>>>>>> accordingly,
>>>>>>>> > > > please don't forget to recompile AMBER.
>>>>>>>> > > > Best Regards
>>>>>>>> > > > Elvis
>>>>>>>> > > >
>>>>>>>> > > >
>>>>>>>> > > >
>>>>>>>> > > > On Mon, 14 Dec 2020 at 17:05, Elvis Martis <
>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>> > > > wrote:
>>>>>>>> > > >
>>>>>>>> > > > > OK.
>>>>>>>> > > > > Have you compiled AMBER20 using the legacy configure method
>>>>>>>> or the
>>>>>>>> > new
>>>>>>>> > > > > cmake method?
>>>>>>>> > > > > If you have using the new cmake method of compilation, then
>>>>>>>> I guess
>>>>>>>> > you
>>>>>>>> > > > > should find that file in
>>>>>>>> > amber20_src/AmberTools/src/sander/mdread2.F90.
>>>>>>>> > > > > if you have compiled using the legacy method, then you
>>>>>>>> files should
>>>>>>>> > in
>>>>>>>> > > > the
>>>>>>>> > > > > path where I sent previously.
>>>>>>>> > > > > Best Regards
>>>>>>>> > > > > Elvis
>>>>>>>> > > > >
>>>>>>>> > > > >
>>>>>>>> > > > >
>>>>>>>> > > > > On Mon, 14 Dec 2020 at 16:55, Vaibhav Dixit <
>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>> > >
>>>>>>>> > > > > wrote:
>>>>>>>> > > > >
>>>>>>>> > > > >> Dear All and Elivs,
>>>>>>>> > > > >> I'm afraid, I still can't find this file inside the
>>>>>>>> AmberTools
>>>>>>>> > folder,
>>>>>>>> > > > >> there is no folder by the same sander.
>>>>>>>> > > > >> All tests passed while installing Amber20, thus I think
>>>>>>>> there is
>>>>>>>> > some
>>>>>>>> > > > >> confusion regarding the directory structure and file
>>>>>>>> locations in
>>>>>>>> > > > Amber18
>>>>>>>> > > > >> and Amber20 as well.
>>>>>>>> > > > >> Please let me know if you know the fix to this as well.
>>>>>>>> > > > >> thank you and best regards.
>>>>>>>> > > > >>
>>>>>>>> > > > >> (base) [exx.c107739 amber20]$ ls
>>>>>>>> > > > >> $AMBERHOME/AmberTools/src/sander/mdread2.F90
>>>>>>>> > > > >> ls: cannot access
>>>>>>>> > > > >>
>>>>>>>> /home/exx/Downloads/amber20/AmberTools/src/sander/mdread2.F90: No
>>>>>>>> > such
>>>>>>>> > > > >> file
>>>>>>>> > > > >> or directory
>>>>>>>> > > > >> (base) [exx.c107739 amber20]$ cd AmberTools/src/
>>>>>>>> > > > >> (base) [exx.c107739 src]$ ls -arlth
>>>>>>>> > > > >> total 28K
>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 cpptraj
>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 mm_pbsa
>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 FEW
>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 pytraj
>>>>>>>> > > > >> drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 ..
>>>>>>>> > > > >> -rw-r--r--. 1 exx exx 1.9K Dec 4 17:26 config.h
>>>>>>>> > > > >> drwxrwxr-x. 6 exx exx 4.0K Dec 4 17:45 .
>>>>>>>> > > > >>
>>>>>>>> > > > >> On Mon, Dec 14, 2020 at 7:46 PM Elvis Martis <
>>>>>>>> > > elvis_bcp.elvismartis.in>
>>>>>>>> > > > >> wrote:
>>>>>>>> > > > >>
>>>>>>>> > > > >> > Hi
>>>>>>>> > > > >> > Sorry my bad. For AMBER20
>>>>>>>> > > > >> > you can look in
>>>>>>>> > > > >> > $AMBERHOME/AmberTools/src/sander/mdread2.F90
>>>>>>>> > > > >> > Start looking from line number 1508 (this should be same
>>>>>>>> unless
>>>>>>>> > > there
>>>>>>>> > > > >> were
>>>>>>>> > > > >> > changes made earlier)
>>>>>>>> > > > >> > this line starts with " else if ( gbsa == 2 ) then"
>>>>>>>> > > > >> >
>>>>>>>> > > > >> > Best Regards
>>>>>>>> > > > >> > Elvis
>>>>>>>> > > > >> >
>>>>>>>> > > > >> >
>>>>>>>> > > > >> >
>>>>>>>> > > > >> > On Mon, 14 Dec 2020 at 15:57, Vaibhav Dixit <
>>>>>>>> > > vaibhavadixit.gmail.com>
>>>>>>>> > > > >> > wrote:
>>>>>>>> > > > >> >
>>>>>>>> > > > >> > > Dear All,
>>>>>>>> > > > >> > > I got this suggestion earlier last month also.
>>>>>>>> > > > >> > > But to my surprise, I can't find these specified
>>>>>>>> folders and
>>>>>>>> > files
>>>>>>>> > > > in
>>>>>>>> > > > >> the
>>>>>>>> > > > >> > > expected locations.
>>>>>>>> > > > >> > > I ran a find command to search for the mdread.f file,
>>>>>>>> but it is
>>>>>>>> > > not
>>>>>>>> > > > >> found
>>>>>>>> > > > >> > > anywhere inside amber20 folder and subfolders.
>>>>>>>> > > > >> > > This command works and finds other files as shown
>>>>>>>> below.
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > Please do let me know what I'm missing to understand
>>>>>>>> here.
>>>>>>>> > > > >> > > thanks
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ ls -arlth
>>>>>>>> > > > >> > > total 128K
>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 7.5K Dec 6 2018 GNU_LGPL_v3
>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 116 Dec 6 2018
>>>>>>>> amber-interactive.sh
>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 1.1K Apr 28 2020 Makefile
>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 37K Apr 28 2020 LICENSE
>>>>>>>> > > > >> > > drwxrwxr-x. 8 exx exx 4.0K Dec 4 15:17 licenses
>>>>>>>> > > > >> > > drwxrwxr-x. 2 exx exx 4.0K Dec 4 15:17 doc
>>>>>>>> > > > >> > > drwxrwxr-x. 19 exx exx 4.0K Dec 4 15:17 miniconda
>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 share
>>>>>>>> > > > >> > > drwxrwxr-x. 9 exx exx 4.0K Dec 4 15:17 benchmarks
>>>>>>>> > > > >> > > drwxrwxr-x. 16 exx exx 4.0K Dec 4 15:17 dat
>>>>>>>> > > > >> > > drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 AmberTools
>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 1.9K Dec 4 17:26 config.h
>>>>>>>> > > > >> > > drwxrwxr-x. 8 exx exx 4.0K Dec 10 15:54 logs
>>>>>>>> > > > >> > > drwxrwxr-x. 157 exx exx 4.0K Dec 10 15:58 test
>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 1.8K Dec 10 20:38 amber.sh
>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 1.9K Dec 10 20:38 amber.csh
>>>>>>>> > > > >> > > drwxrwxr-x. 15 exx exx 4.0K Dec 10 21:50 .
>>>>>>>> > > > >> > > drwxr-xr-x. 2 exx exx 4.0K Dec 10 21:50 include
>>>>>>>> > > > >> > > drwxrwxr-x. 4 exx exx 4.0K Dec 10 21:50 lib64
>>>>>>>> > > > >> > > drwxr-xr-x. 4 exx exx 4.0K Dec 10 21:50 lib
>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 10 21:50 bin
>>>>>>>> > > > >> > > drwxr-xr-x. 24 exx exx 4.0K Dec 12 12:13 ..
>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ cd AmberTools/
>>>>>>>> > > > >> > > (base) [exx.c107739 AmberTools]$ ls -arlth
>>>>>>>> > > > >> > > total 28K
>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 benchmarks
>>>>>>>> > > > >> > > drwxrwxr-x. 6 exx exx 4.0K Dec 4 15:17 examples
>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:25 .pytest_cache
>>>>>>>> > > > >> > > drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 .
>>>>>>>> > > > >> > > drwxrwxr-x. 6 exx exx 4.0K Dec 4 17:45 src
>>>>>>>> > > > >> > > drwxrwxr-x. 15 exx exx 4.0K Dec 10 21:50 ..
>>>>>>>> > > > >> > > drwxrwxr-x. 65 exx exx 4.0K Dec 10 21:59 test
>>>>>>>> > > > >> > > (base) [exx.c107739 AmberTools]$ cd ..
>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name mdread.f
>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name amber.sh
>>>>>>>> > > > >> > > ./amber.sh
>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name am1bcc
>>>>>>>> > > > >> > > ./bin/am1bcc
>>>>>>>> > > > >> > > ./bin/wrapped_progs/am1bcc
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > On Mon, Dec 14, 2020 at 7:14 PM Elvis Martis <
>>>>>>>> > > > >> elvis_bcp.elvismartis.in>
>>>>>>>> > > > >> > > wrote:
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > > Hi
>>>>>>>> > > > >> > > > Here is a solution in a different thread, you will
>>>>>>>> have to
>>>>>>>> > edit
>>>>>>>> > > > the
>>>>>>>> > > > >> > > > mdread.f file. Please have a look at link below to
>>>>>>>> know how to
>>>>>>>> > > > edit
>>>>>>>> > > > >> the
>>>>>>>> > > > >> > > > file and where to find it.
>>>>>>>> > > > >> > > > Re: [AMBER] bad atom type: Br from Jason Swails on
>>>>>>>> 2011-11-11
>>>>>>>> > > > (Amber
>>>>>>>> > > > >> > > > Archive Nov 2011) (ambermd.org)
>>>>>>>> > > > >> > > > <http://archive.ambermd.org/201111/0370.html>
>>>>>>>> > > > >> > > > Best Regards
>>>>>>>> > > > >> > > > Elvis
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > > > On Mon, 14 Dec 2020 at 15:34, Vaibhav Dixit <
>>>>>>>> > > > >> vaibhavadixit.gmail.com>
>>>>>>>> > > > >> > > > wrote:
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > > > > Dear All,
>>>>>>>> > > > >> > > > > I'm getting the following error message while
>>>>>>>> trying to run
>>>>>>>> > > > >> MMPBSA.py
>>>>>>>> > > > >> > > in
>>>>>>>> > > > >> > > > > Amber20.
>>>>>>>> > > > >> > > > > It says that the receptor mask overwritten with
>>>>>>>> default,
>>>>>>>> > > which I
>>>>>>>> > > > >> > don't
>>>>>>>> > > > >> > > > know
>>>>>>>> > > > >> > > > > why it is doing even after I gave a specific
>>>>>>>> receptor mask
>>>>>>>> > > (1st
>>>>>>>> > > > >> > residue
>>>>>>>> > > > >> > > > is
>>>>>>>> > > > >> > > > > my ligand in the trajectory). It also gives the
>>>>>>>> bad atom
>>>>>>>> > type
>>>>>>>> > > > >> error,
>>>>>>>> > > > >> > > > that
>>>>>>>> > > > >> > > > > I got with Amber18 last month.
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > >From the output files (attached), it is not clear
>>>>>>>> what is
>>>>>>>> > the
>>>>>>>> > > > >> exact
>>>>>>>> > > > >> > > > nature
>>>>>>>> > > > >> > > > > of the error, thus I have no clue how to proceed
>>>>>>>> to fix the
>>>>>>>> > > > same.
>>>>>>>> > > > >> > > > > Can you please help me understand why the dry
>>>>>>>> prmtop is not
>>>>>>>> > > > >> > compatible?
>>>>>>>> > > > >> > > > > And how can I possibly attempt to get this
>>>>>>>> analysis done?
>>>>>>>> > > > >> > > > > My mmpbsa input file is given below.
>>>>>>>> > > > >> > > > > Thanking you all in advance.
>>>>>>>> > > > >> > > > > Best regards
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > mmpbsa input file:
>>>>>>>> > > > >> > > > > (base) [exx.c107739 mmpbsa-6701-tyr]$ more
>>>>>>>> mmpbsa-decomp.in
>>>>>>>> > > > >> > > > > &general
>>>>>>>> > > > >> > > > > startframe=1, endframe=100, interval=5,
>>>>>>>> > > > >> > > > > verbose=2, keep_files=2, netcdf=1,
>>>>>>>> receptor_mask=2-386,
>>>>>>>> > > > >> > > > > strip_mask=:387-16750,
>>>>>>>> > > > >> > > > > /
>>>>>>>> > > > >> > > > > &gb
>>>>>>>> > > > >> > > > > igb=5, saltcon=0.150,
>>>>>>>> > > > >> > > > > /
>>>>>>>> > > > >> > > > > &pb
>>>>>>>> > > > >> > > > > istrng=0.15, fillratio=4.0,
>>>>>>>> > > > >> > > > > /
>>>>>>>> > > > >> > > > > &decomp
>>>>>>>> > > > >> > > > > idecomp=1, print_res="56-57; 74-75; 167; 222;
>>>>>>>> 225-227;
>>>>>>>> > > > 229-231;
>>>>>>>> > > > >> > > > 272-273;
>>>>>>>> > > > >> > > > > 275-277; 336; 368; 370; 372-373"
>>>>>>>> > > > >> > > > > dec_verbose=1,
>>>>>>>> > > > >> > > > > /
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Error message on the terminal
>>>>>>>> > > > >> > > > > (base) [exx.c107739 mmpbsa-6701-tyr]$ MMPBSA.py
>>>>>>>> -O -i
>>>>>>>> > > > >> > mmpbsa-decomp.in
>>>>>>>> > > > >> > > > -o
>>>>>>>> > > > >> > > > > mmpbs-rouf-6701-tyr.dat -do
>>>>>>>> decomp-rouf-6701-tyr.dat -sp
>>>>>>>> > > > >> > > > > ../rouf-6701-tyr-solv.prmtop -cp
>>>>>>>> ../rouf-6701-tyr-dry.prmtop
>>>>>>>> > > -lp
>>>>>>>> > > > >> > > > > ../tyr-dry.prmtop -y ../
>>>>>>>> rouf-6701-tyr-solv-prod300.nc -rp
>>>>>>>> > > > >> > > > > ../rouf-6701-nolig-dry.prmtop
>>>>>>>> > > > >> > > > > Loading and checking parameter files for
>>>>>>>> compatibility...
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> >
>>>>>>>> > > > >>
>>>>>>>> > > >
>>>>>>>> > >
>>>>>>>> >
>>>>>>>> */home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py:603:
>>>>>>>> > > > >> > > > > UserWarning: receptor_mask overwritten with
>>>>>>>> default*
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > warnings.warn('receptor_mask overwritten with
>>>>>>>> default\n')
>>>>>>>> > > > >> > > > > cpptraj found! Using
>>>>>>>> /home/exx/Downloads/amber20/bin/cpptraj
>>>>>>>> > > > >> > > > > sander found! Using
>>>>>>>> /home/exx/Downloads/amber20/bin/sander
>>>>>>>> > > > >> > > > > Preparing trajectories for simulation...
>>>>>>>> > > > >> > > > > 20 frames were processed by cpptraj for use in
>>>>>>>> calculation.
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Running calculations on normal system...
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Beginning GB calculations with
>>>>>>>> > > > >> /home/exx/Downloads/amber20/bin/sander
>>>>>>>> > > > >> > > > > calculating complex contribution...
>>>>>>>> > > > >> > > > > * bad atom type: M1*
>>>>>>>> > > > >> > > > > File
>>>>>>>> "/home/exx/Downloads/amber20/bin/MMPBSA.py", line
>>>>>>>> > 100,
>>>>>>>> > > in
>>>>>>>> > > > >> > > <module>
>>>>>>>> > > > >> > > > > app.run_mmpbsa()
>>>>>>>> > > > >> > > > > File
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> >
>>>>>>>> > > > >>
>>>>>>>> > > >
>>>>>>>> > >
>>>>>>>> >
>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py",
>>>>>>>> > > > >> > > > > line 218, in run_mmpbsa
>>>>>>>> > > > >> > > > > self.calc_list.run(rank, self.stdout)
>>>>>>>> > > > >> > > > > File
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> >
>>>>>>>> > > > >>
>>>>>>>> > > >
>>>>>>>> > >
>>>>>>>> >
>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>>>>>>>> > > > >> > > > > line 82, in run
>>>>>>>> > > > >> > > > > calc.run(rank, stdout=stdout, stderr=stderr)
>>>>>>>> > > > >> > > > > File
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> >
>>>>>>>> > > > >>
>>>>>>>> > > >
>>>>>>>> > >
>>>>>>>> >
>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>>>>>>>> > > > >> > > > > line 156, in run
>>>>>>>> > > > >> > > > > raise CalcError('%s failed with prmtop %s!' %
>>>>>>>> > > (self.program,
>>>>>>>> > > > >> > > > > CalcError: /home/exx/Downloads/amber20/bin/sander
>>>>>>>> failed
>>>>>>>> > with
>>>>>>>> > > > >> prmtop
>>>>>>>> > > > >> > > > > ../rouf-6701-tyr-dry.prmtop!
>>>>>>>> > > > >> > > > > Exiting. All files have been retained.
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > --
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Regards,
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Dr. Vaibhav A. Dixit,
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > Visiting Scientist at the Manchester Institute of
>>>>>>>> > > Biotechnology
>>>>>>>> > > > >> > (MIB),
>>>>>>>> > > > >> > > > The
>>>>>>>> > > > >> > > > > University of Manchester, 131 Princess Street,
>>>>>>>> Manchester M1
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> > > > 7DN,
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> > > > >> UK
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> .
>>>>>>>> > > > >> > > > > AND
>>>>>>>> > > > >> > > > > Assistant Professor,
>>>>>>>> > > > >> > > > > Department of Pharmacy,
>>>>>>>> > > > >> > > > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>> > > > >> > > > > Birla Institute of Technology and Sciences Pilani
>>>>>>>> > > (BITS-Pilani),
>>>>>>>> > > > >> > > > > VidyaVihar Campus, street number 41, Pilani,
>>>>>>>> Rajasthan
>>>>>>>> > 333031.
>>>>>>>> > > > >> > > > > India.
>>>>>>>> > > > >> > > > > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>> > > > >> > > > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>> > > > >> > vaibhavadixit.gmail.com
>>>>>>>> > > > >> > > > >
>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>> > > > >> > > > >
>>>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >>
>>>>>>>> >
>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > > P Please consider the environment before printing
>>>>>>>> this
>>>>>>>> > > > >> > > > > _______________________________________________
>>>>>>>> > > > >> > > > > AMBER mailing list
>>>>>>>> > > > >> > > > > AMBER.ambermd.org
>>>>>>>> > > > >> > > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > > >> > > > >
>>>>>>>> > > > >> > > > _______________________________________________
>>>>>>>> > > > >> > > > AMBER mailing list
>>>>>>>> > > > >> > > > AMBER.ambermd.org
>>>>>>>> > > > >> > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > > >> > > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > --
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > Regards,
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > Dr. Vaibhav A. Dixit,
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > Visiting Scientist at the Manchester Institute of
>>>>>>>> Biotechnology
>>>>>>>> > > > (MIB),
>>>>>>>> > > > >> > The
>>>>>>>> > > > >> > > University of Manchester, 131 Princess Street,
>>>>>>>> Manchester M1
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> > 7DN,
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> > > > UK
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> .
>>>>>>>> > > > >> > > AND
>>>>>>>> > > > >> > > Assistant Professor,
>>>>>>>> > > > >> > > Department of Pharmacy,
>>>>>>>> > > > >> > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>> > > > >> > > Birla Institute of Technology and Sciences Pilani
>>>>>>>> (BITS-Pilani),
>>>>>>>> > > > >> > > VidyaVihar Campus, street number 41, Pilani, Rajasthan
>>>>>>>> 333031.
>>>>>>>> > > > >> > > India.
>>>>>>>> > > > >> > > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>> > > > >> > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>> > > > >> vaibhavadixit.gmail.com
>>>>>>>> > > > >> > >
>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>> > > > >> > > https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > >
>>>>>>>> > > >
>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > > P Please consider the environment before printing this
>>>>>>>> > > > >> > > _______________________________________________
>>>>>>>> > > > >> > > AMBER mailing list
>>>>>>>> > > > >> > > AMBER.ambermd.org
>>>>>>>> > > > >> > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > > >> > >
>>>>>>>> > > > >> > _______________________________________________
>>>>>>>> > > > >> > AMBER mailing list
>>>>>>>> > > > >> > AMBER.ambermd.org
>>>>>>>> > > > >> > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > > >> >
>>>>>>>> > > > >>
>>>>>>>> > > > >>
>>>>>>>> > > > >> --
>>>>>>>> > > > >>
>>>>>>>> > > > >> Regards,
>>>>>>>> > > > >>
>>>>>>>> > > > >> Dr. Vaibhav A. Dixit,
>>>>>>>> > > > >>
>>>>>>>> > > > >> Visiting Scientist at the Manchester Institute of
>>>>>>>> Biotechnology
>>>>>>>> > (MIB),
>>>>>>>> > > > The
>>>>>>>> > > > >> University of Manchester, 131 Princess Street, Manchester
>>>>>>>> M1 7DN,
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> > UK
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>> .
>>>>>>>> > > > >> AND
>>>>>>>> > > > >> Assistant Professor,
>>>>>>>> > > > >> Department of Pharmacy,
>>>>>>>> > > > >> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>> > > > >> Birla Institute of Technology and Sciences Pilani
>>>>>>>> (BITS-Pilani),
>>>>>>>> > > > >> VidyaVihar Campus, street number 41, Pilani, Rajasthan
>>>>>>>> 333031.
>>>>>>>> > > > >> India.
>>>>>>>> > > > >> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>> > > > >> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>> > > vaibhavadixit.gmail.com
>>>>>>>> > > > >> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>> > > > >> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>> > > > >>
>>>>>>>> > > > >> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>> > > > >>
>>>>>>>> > > > >>
>>>>>>>> >
>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>> > > > >>
>>>>>>>> > > > >> P Please consider the environment before printing this
>>>>>>>> > > > >> _______________________________________________
>>>>>>>> > > > >> AMBER mailing list
>>>>>>>> > > > >> AMBER.ambermd.org
>>>>>>>> > > > >> http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > > >>
>>>>>>>> > > > >
>>>>>>>> > > > _______________________________________________
>>>>>>>> > > > AMBER mailing list
>>>>>>>> > > > AMBER.ambermd.org
>>>>>>>> > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > > >
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> > > --
>>>>>>>> > >
>>>>>>>> > > Regards,
>>>>>>>> > >
>>>>>>>> > > Dr. Vaibhav A. Dixit,
>>>>>>>> > >
>>>>>>>> > > Visiting Scientist at the Manchester Institute of Biotechnology
>>>>>>>> (MIB),
>>>>>>>> > The
>>>>>>>> > > University of Manchester, 131 Princess Street, Manchester M1
>>>>>>>> 7DN, UK
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>>>> .
>>>>>>>> > > AND
>>>>>>>> > > Assistant Professor,
>>>>>>>> > > Department of Pharmacy,
>>>>>>>> > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>> > > Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>>>>> > > VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>>>> > > India.
>>>>>>>> > > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>> > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>> > > http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>> > > https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>> > >
>>>>>>>> > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>> > >
>>>>>>>> > >
>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>> > >
>>>>>>>> > > P Please consider the environment before printing this e-mail
>>>>>>>> > > _______________________________________________
>>>>>>>> > > AMBER mailing list
>>>>>>>> > > AMBER.ambermd.org
>>>>>>>> > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> > >
>>>>>>>> > _______________________________________________
>>>>>>>> > AMBER mailing list
>>>>>>>> > AMBER.ambermd.org
>>>>>>>> > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>> >
>>>>>>>>
>>>>>>>>
>>>>>>>> --
>>>>>>>>
>>>>>>>> Regards,
>>>>>>>>
>>>>>>>> Dr. Vaibhav A. Dixit,
>>>>>>>>
>>>>>>>> Visiting Scientist at the Manchester Institute of Biotechnology
>>>>>>>> (MIB), The
>>>>>>>> University of Manchester, 131 Princess Street, Manchester M1 7DN,
>>>>>>>> UK
>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>>>> .
>>>>>>>> AND
>>>>>>>> Assistant Professor,
>>>>>>>> Department of Pharmacy,
>>>>>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>>>> India.
>>>>>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>
>>>>>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>
>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>
>>>>>>>> P Please consider the environment before printing this e-mail
>>>>>>>> _______________________________________________
>>>>>>>> AMBER mailing list
>>>>>>>> AMBER.ambermd.org
>>>>>>>> http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>
>>>>>>>
>>>>>>
>>>>>> --
>>>>>>
>>>>>> Regards,
>>>>>>
>>>>>> Dr. Vaibhav A. Dixit,
>>>>>>
>>>>>> Visiting Scientist at the Manchester Institute of Biotechnology
>>>>>> (MIB), The University of Manchester, 131 Princess Street, Manchester
>>>>>> M1 7DN, UK
>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>> .
>>>>>> AND
>>>>>> Assistant Professor,
>>>>>> Department of Pharmacy,
>>>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>> India.
>>>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>> vaibhavadixit.gmail.com
>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>
>>>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>
>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>
>>>>>> P Please consider the environment before printing this e-mail
>>>>>>
>>>>>>
>>>>
>>>> --
>>>>
>>>> Regards,
>>>>
>>>> Dr. Vaibhav A. Dixit,
>>>>
>>>> Visiting Scientist at the Manchester Institute of Biotechnology (MIB),
>>>> The University of Manchester, 131 Princess Street, Manchester M1 7DN,
>>>> UK
>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>> .
>>>> AND
>>>> Assistant Professor,
>>>> Department of Pharmacy,
>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>> India.
>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>
>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>
>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>
>>>> P Please consider the environment before printing this e-mail
>>>>
>>>>
>>>
>>> --
>>> Best Regards
>>> Elvis
>>>
>>
>
> --
>
> Regards,
>
> Dr. Vaibhav A. Dixit,
>
> Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The
> University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
> AND
> Assistant Professor,
> Department of Pharmacy,
> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
> India.
> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>
> ORCID ID: https://orcid.org/0000-0003-4015-2941
>
> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>
> P Please consider the environment before printing this e-mail
>
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: 02-image.png)
