Date: Tue, 15 Dec 2020 14:38:04 +0900
Dear Elvis and All,
The PB calculation seems to be running fine now with the radiopt=0 option.
I'm trying with the serial version/mode. Once the serial jobs finish
correctly, then I will recompile, test the Amber parallel version and
resubmit these calculations to check for speed up gained.
Is it possible to estimate how much extra time the PB calculations are
likely to take than the GB? i.e. what is the expected scaling factor
between GB and PB calculations? For 20 frames the GB calculation took
about 30 minutes.
As I understand from the manual, MMPBSA runs only on CPU, please let me
know if GPU usage is possible.
Thank you very much for your valuable suggestions and support.
Best regards.
On Tue, Dec 15, 2020 at 4:33 PM Elvis Martis <elvis_bcp.elvismartis.in>
wrote:
> Can you confirm if you are using radiopt=0 (default is =1) for PB
> calculation?
> Also, try to run mmpbsa.py in serial mode to check if it runs without
> errors/.
> Best Regards
> Elvis
>
>
>
> On Tue, 15 Dec 2020 at 12:56, Vaibhav Dixit <vaibhavadixit.gmail.com>
> wrote:
>
>> Dear All and Elvis,
>> The GB calculation did work after recompling with the suggested changes.
>> But the PB calculation is failing with the following error messages.
>> Inspection of the output file shows that it did not find radii for the S
>> atom which has type "Y1" as defined by the MCPB.py tool which I had used
>> for building parameters for the co-factor.
>> Thus I'm wondering does this require adding similar lines in the PB
>> section of the mdread2.F90 file?
>> Please help me understand how should I try to fix this error?
>> Thank you and best regards.
>>
>> tail of the _MMPBSA_complex_pb.mdout.0 file
>> Frozen or restrained atoms:
>> ibelly = 0, ntr = 0
>>
>> Energy minimization:
>> maxcyc = 1, ncyc = 10, ntmin = 1
>> dx0 = 0.01000, drms = 0.00010
>> PB Bomb in pb_aaradi(): No radius assigned for atom 5179 SG Y1
>>
>> (base) [exx.c107739 mmpbsa-6701-tyr]$ MMPBSA.py -O -i mmpbsa-decomp.in
>> -o mmpbs-rouf-6701-tyr.dat -do decomp-rouf-6701-tyr.dat -sp
>> ../rouf-6701-tyr-solv.prmtop -cp ../rouf-6701-tyr-dry.prmtop -lp
>> ../tyr-dry.prmtop -y ../rouf-6701-tyr-solv-prod300.nc -rp
>> ../rouf-6701-nolig-dry.prmtop
>> Loading and checking parameter files for compatibility...
>> /home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py:603:
>> UserWarning: receptor_mask overwritten with default
>>
>> warnings.warn('receptor_mask overwritten with default\n')
>> cpptraj found! Using /home/exx/Downloads/amber20/bin/cpptraj
>> sander found! Using /home/exx/Downloads/amber20/bin/sander
>> Preparing trajectories for simulation...
>> 20 frames were processed by cpptraj for use in calculation.
>>
>> Running calculations on normal system...
>>
>> Beginning GB calculations with /home/exx/Downloads/amber20/bin/sander
>> calculating complex contribution...
>> calculating receptor contribution...
>> calculating ligand contribution...
>>
>> Beginning PB calculations with /home/exx/Downloads/amber20/bin/sander
>> calculating complex contribution...
>> File "/home/exx/Downloads/amber20/bin/MMPBSA.py", line 100, in <module>
>> app.run_mmpbsa()
>> File
>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py",
>> line 218, in run_mmpbsa
>> self.calc_list.run(rank, self.stdout)
>> File
>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>> line 82, in run
>> calc.run(rank, stdout=stdout, stderr=stderr)
>> File
>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>> line 428, in run
>> error_list = [s.strip() for s in out.split('\n')
>> TypeError: a bytes-like object is required, not 'str'
>> Exiting. All files have been retained.
>>
>>
>>
>> On Tue, Dec 15, 2020 at 2:39 AM Elvis Martis <elvis_bcp.elvismartis.in>
>> wrote:
>>
>>> Hi Vaibhav
>>> I could reproduce the error.
>>> The reason for it is, I edited the mdread2.F90 file using notepad++.
>>> Apparently, the UTC-8 Unicode in windows has some issues and seems
>>> incompatible with Linux, and therefore, the unknown characters.
>>>
>>> I would recommend, you do a fresh install, untar the AmberTools20.bz2
>>> file and go to AmberTools/src/sander and add (i suggest typing it) the
>>> required lines in the mdread2.F90 using any editor in Linux itself.
>>> It should solve the problem, as I could compile it.
>>> Best Regards
>>> Elvis
>>>
>>>
>>>
>>> On Mon, 14 Dec 2020 at 19:55, Elvis Martis <elvis_bcp.elvismartis.in>
>>> wrote:
>>>
>>>> I will try compile amber20 Using this very mdread2.F90 file and try to
>>>> reproduce this error.
>>>> I will get back to you asap
>>>>
>>>> On Monday, December 14, 2020, Vaibhav Dixit <vaibhavadixit.gmail.com>
>>>> wrote:
>>>>
>>>>> Dear Elivs,
>>>>> This file is giving me the same error.
>>>>> I'm not sure if it is related to the file getting transferred over the
>>>>> internet which might lead to strange characters and sort of things.
>>>>> Please do let me know if you find a way to fix that.
>>>>> thanks
>>>>>
>>>>> (base) [exx.c107739 build]$ make install
>>>>> [ 1%] Built target ucpp
>>>>> [ 1%] Built target dft_scalar
>>>>> [ 1%] Built target fftw3_threads_obj
>>>>> [ 1%] Built target reodft
>>>>> [ 1%] Built target simd_support
>>>>> [ 1%] Built target rdft_scalar_r2r
>>>>> [ 1%] Built target rdft_sse2_codelets
>>>>> [ 2%] Built target fftw_kernel
>>>>> [ 2%] Built target rdft_scalar
>>>>> [ 5%] Built target rdft_scalar_r2cb
>>>>> [ 7%] Built target rdft_scalar_r2cf
>>>>> [ 8%] Built target dft_scalar_codelets
>>>>> [ 12%] Built target dft_sse2_codelets
>>>>> [ 13%] Built target rdft
>>>>> [ 15%] Built target fftw_api
>>>>> [ 16%] Built target dft
>>>>> [ 16%] Built target fftw
>>>>> [ 17%] Built target libfftw_bench
>>>>> [ 17%] Built target fftw_benchmark_common_obj
>>>>> [ 17%] Built target fftw_benchmark
>>>>> [ 17%] Built target fftw_wisdom
>>>>> [ 18%] Built target dispatch
>>>>> [ 18%] Built target netcdf3
>>>>> [ 18%] Built target netcdf
>>>>> [ 18%] Built target ncgen
>>>>> [ 18%] Built target ncgen3
>>>>> [ 18%] Built target nccopy
>>>>> [ 19%] Built target ncdump
>>>>> [ 20%] Built target netcdff
>>>>> [ 20%] Built target xblas_build
>>>>> [ 22%] Built target blas
>>>>> [ 28%] Built target lapack
>>>>> [ 29%] Built target arpack
>>>>> [ 29%] Built target boost_build
>>>>> [ 29%] Built target amber_common
>>>>> [ 30%] Built target libpbsa
>>>>> [ 30%] Built target sff_lex
>>>>> [ 30%] Built target dsarpack_obj
>>>>> [ 32%] Built target rism
>>>>> [ 32%] Built target sff_rism_interface
>>>>> [ 32%] Built target sff
>>>>> [ 33%] Built target gbnsr6
>>>>> [ 33%] Built target cifparse
>>>>> [ 34%] Built target addles
>>>>> [ 36%] Built target sqm_common
>>>>> [ 36%] Built target libsqm
>>>>> [ 37%] Built target sff_fortran
>>>>> [ 37%] Built target sander_rism_interface
>>>>> Scanning dependencies of target sander_base_obj
>>>>> make[2]: Warning: File `../AmberTools/src/sander/mdread2.F90' has
>>>>> modification time 12412 s in the future
>>>>> [ 37%] Building Fortran object
>>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1567.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> else if (atomicnumber .eq. 26) then
>>>>> 1
>>>>> Warning: Nonconforming tab character at (1)
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1568.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 !
>>>>> \xC2\xA0Fe ra
>>>>> 1
>>>>> Error: Invalid character in name at (1)
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1569.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>> x(L165-1+i) = 1
>>>>> 1
>>>>> Error: Invalid character in name at (1)
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1630.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> else if (atomicnumber .eq. 26) then
>>>>> 1
>>>>> Warning: Nonconforming tab character at (1)
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1631.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 ! \xC2\xA0Fe
>>>>> radius = 1.3
>>>>> 1
>>>>> Error: Invalid character in name at (1)
>>>>>
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1632.1:
>>>>> Included at
>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>
>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 x(L165-1+i) =
>>>>> 1.39d0 + 1.
>>>>> 1
>>>>> Error: Invalid character in name at (1)
>>>>> make[2]: ***
>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o] Error 1
>>>>> make[1]: ***
>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all] Error 2
>>>>> make: *** [all] Error 2
>>>>> (base) [exx.c107739 build]$
>>>>>
>>>>> On Mon, Dec 14, 2020 at 7:30 PM Elvis Martis <elvis_bcp.elvismartis.in>
>>>>> wrote:
>>>>>
>>>>>> Hi Vaibhav
>>>>>> The problem was that tabs and indentations were not correct.
>>>>>> Also, the x(L165-1+i) = 1.386d0 + 1.4d0 was changed to x(L165-1+i)
>>>>>> = 1.39d0 + 1.4d0
>>>>>> This time I made sure they are in the right place.
>>>>>> I hope this time it will OK
>>>>>> Best Regards
>>>>>> Elvis
>>>>>>
>>>>>>
>>>>>>
>>>>>> On Mon, 14 Dec 2020 at 19:13, Vaibhav Dixit <vaibhavadixit.gmail.com>
>>>>>> wrote:
>>>>>>
>>>>>>> Dear Elvis,
>>>>>>> I have copied your file in the sander folder and reran the run_cmake
>>>>>>> and make install commands.
>>>>>>> But I'm getting new errors after a few steps shown below.
>>>>>>> I think it still doesn't like the modifications and syntax like
>>>>>>> errors are probably showing up.
>>>>>>> Please let me know what modifications should I make now.
>>>>>>> thanks for your continued support and suggestions.
>>>>>>> Best regards.
>>>>>>>
>>>>>>> Scanning dependencies of target sander_base_obj
>>>>>>> [ 37%] Building Fortran object
>>>>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1567.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> else if (atomicnumber .eq. 26) then
>>>>>>> 1
>>>>>>> Warning: Nonconforming tab character at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1568.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0!
>>>>>>> \xC2\xA0Fe rad
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1569.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>> x(L165-1+i) = 1
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1570.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 else
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1571.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>> \xC2\xA0write( 0
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1572.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>> \xC2\xA0call mex
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1630.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> else if (atomicnumber .eq. 26) then
>>>>>>> 1
>>>>>>> Warning: Nonconforming tab character at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1631.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0!
>>>>>>> \xC2\xA0Fe rad
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>>
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1632.1:
>>>>>>> Included at
>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>
>>>>>>> \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>> x(L165-1+i) = 1
>>>>>>> 1
>>>>>>> Error: Invalid character in name at (1)
>>>>>>> make[2]: ***
>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o] Error 1
>>>>>>> make[1]: ***
>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all] Error 2
>>>>>>> make: *** [all] Error 2
>>>>>>>
>>>>>>> On Mon, Dec 14, 2020 at 10:31 PM Elvis Martis <
>>>>>>> elvis_bcp.elvismartis.in> wrote:
>>>>>>>
>>>>>>>> Hi Vaibhav
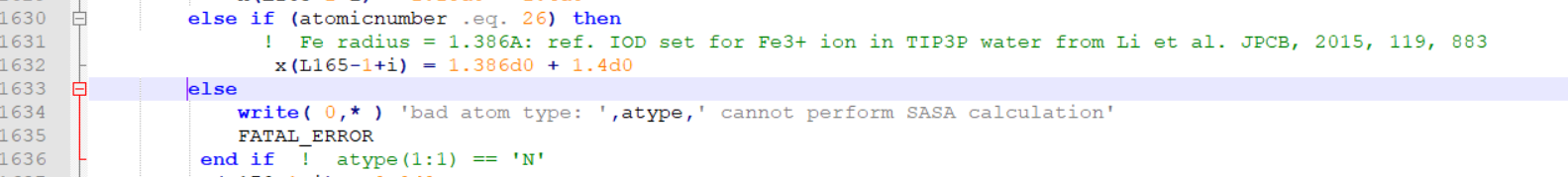
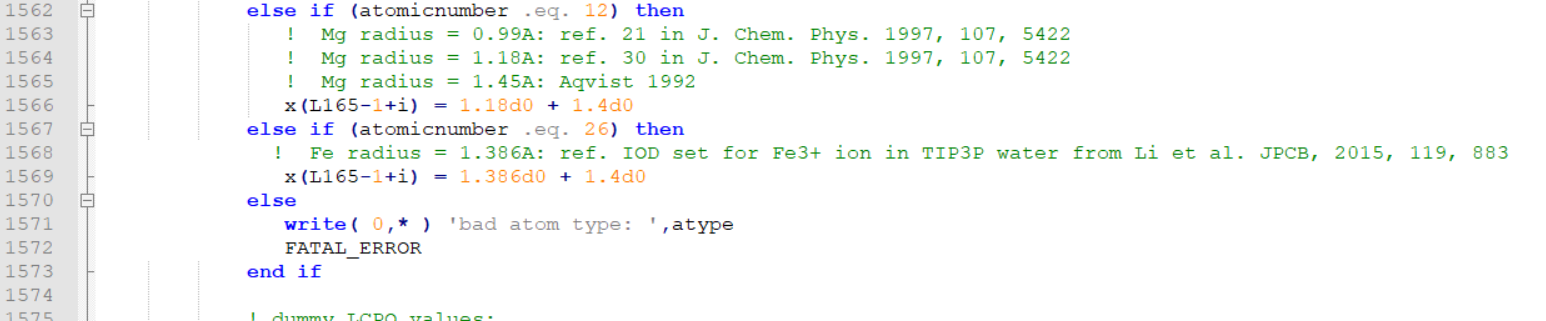
>>>>>>>> I guess the error was because of the commenting the "else" line.
>>>>>>>> I have edited mdread2.F90 and attached here.
>>>>>>>> I am pasting the snapshots of the changes I did at two places.
>>>>>>>> [image: image.png]
>>>>>>>>
>>>>>>>> [image: image.png]
>>>>>>>>
>>>>>>>> You can directly paste this mread2.F90 in the src/sander and
>>>>>>>> recompile.
>>>>>>>> Best Regards
>>>>>>>> Elvis
>>>>>>>>
>>>>>>>>
>>>>>>>>
>>>>>>>> On Mon, 14 Dec 2020 at 18:32, Vaibhav Dixit <
>>>>>>>> vaibhavadixit.gmail.com> wrote:
>>>>>>>>
>>>>>>>>> Dear Elvis,
>>>>>>>>> I searched for the igb == 2 and then "bad atom type".
>>>>>>>>> I commented out the following lines (as shown).
>>>>>>>>> these are line number 1567, 1568 and 1569 in the attached file.
>>>>>>>>>
>>>>>>>>> ! else
>>>>>>>>> ! write( 0,* ) 'bad atom type: ',atype
>>>>>>>>> ! FATAL_ERROR
>>>>>>>>>
>>>>>>>>> Then added the following lines
>>>>>>>>> else if (atomicnumber .eq. 23) then
>>>>>>>>> ! Fe radius = 1.386A: ref. IOD set for Fe3+ ion in
>>>>>>>>> TIP3P
>>>>>>>>> water from Li et al. JPCB, 2015, 119, 883
>>>>>>>>> x(L165-1+i) = 1.386d0 + 1.4d0
>>>>>>>>>
>>>>>>>>> These line numbers are 1570, 1571 and 1572 in the attached file.
>>>>>>>>>
>>>>>>>>> Please let me know if I need to make additional changes in that
>>>>>>>>> file.
>>>>>>>>> Thank you and best regards.
>>>>>>>>>
>>>>>>>>>
>>>>>>>>> On Mon, Dec 14, 2020 at 9:53 PM Elvis Martis <
>>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>>> wrote:
>>>>>>>>>
>>>>>>>>> > Can you point out what changes you did exactly?
>>>>>>>>> >
>>>>>>>>> >
>>>>>>>>> > Best Regards
>>>>>>>>> > Elvis
>>>>>>>>> >
>>>>>>>>> >
>>>>>>>>> >
>>>>>>>>> > On Mon, 14 Dec 2020 at 18:14, Vaibhav Dixit <
>>>>>>>>> vaibhavadixit.gmail.com>
>>>>>>>>> > wrote:
>>>>>>>>> >
>>>>>>>>> > > Dear All and Elivs,
>>>>>>>>> > > OK, now I got the file in amber20_src/AmberTools/src/sander.
>>>>>>>>> > > I have made the following changes (in bold), and then set DMPI
>>>>>>>>> and DCUDA
>>>>>>>>> > > both =FALSE and then ran run_make script.
>>>>>>>>> > > But I get the error shown at the end.
>>>>>>>>> > > Can you please check and help me understand what corrections
>>>>>>>>> to me to
>>>>>>>>> > these
>>>>>>>>> > > lines?
>>>>>>>>> > >
>>>>>>>>> > > Thanks a lot and best regards.
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> > > x(L165-1+i) = 1.10d0 + 1.4d0
>>>>>>>>> > > else if (atomicnumber .eq. 12) then
>>>>>>>>> > > ! Mg radius = 0.99A: ref. 21 in J. Chem. Phys.
>>>>>>>>> 1997, 107,
>>>>>>>>> > > 5422
>>>>>>>>> > > ! Mg radius = 1.18A: ref. 30 in J. Chem. Phys.
>>>>>>>>> 1997, 107,
>>>>>>>>> > > 5422
>>>>>>>>> > > ! Mg radius = 1.45A: Aqvist 1992
>>>>>>>>> > > x(L165-1+i) = 1.18d0 + 1.4d0
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> > > *# else# write( 0,* ) 'bad atom type:
>>>>>>>>> ',atype#
>>>>>>>>> > > FATAL_ERROR*
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> > > * else if (atomicnumber .eq. 23) then
>>>>>>>>> ! Fe
>>>>>>>>> > radius
>>>>>>>>> > > = 1.386A: ref. IOD set for Fe3+ ion in TIP3P water from Li et
>>>>>>>>> al. JPCB,
>>>>>>>>> > > 2015, 119, 883 x(L165-1+i) = 1.386d0 + 1.4d0*
>>>>>>>>> > > end if
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> > > *Error with run_cmake script*
>>>>>>>>> > > (base) [exx.c107739 build]$ gedit run_cmake &
>>>>>>>>> > > [2] 16544
>>>>>>>>> > > (base) [exx.c107739 build]$ make install
>>>>>>>>> > > [ 0%] Built target ucpp
>>>>>>>>> > > [ 0%] Built target fftw3_threads_obj
>>>>>>>>> > > [ 1%] Built target fftw_api
>>>>>>>>> > > [ 1%] Built target dft
>>>>>>>>> > > [ 1%] Built target dft_scalar
>>>>>>>>> > > [ 2%] Built target dft_scalar_codelets
>>>>>>>>> > > [ 5%] Built target dft_sse2_codelets
>>>>>>>>> > > [ 5%] Built target rdft
>>>>>>>>> > > [ 5%] Built target rdft_scalar
>>>>>>>>> > > [ 7%] Built target rdft_scalar_r2cb
>>>>>>>>> > > [ 8%] Built target rdft_scalar_r2cf
>>>>>>>>> > > [ 8%] Built target rdft_scalar_r2r
>>>>>>>>> > > [ 8%] Built target rdft_sse2_codelets
>>>>>>>>> > > [ 9%] Built target fftw_kernel
>>>>>>>>> > > [ 9%] Built target reodft
>>>>>>>>> > > [ 9%] Built target simd_support
>>>>>>>>> > > [ 9%] Built target fftw
>>>>>>>>> > > [ 9%] Built target libfftw_bench
>>>>>>>>> > > [ 9%] Built target fftw_benchmark_common_obj
>>>>>>>>> > > [ 9%] Built target fftw_benchmark
>>>>>>>>> > > [ 9%] Built target fftw_wisdom
>>>>>>>>> > > [ 9%] Built target fftw_mpi
>>>>>>>>> > > [ 9%] Built target fftw_mpi_benchmark
>>>>>>>>> > > [ 9%] Built target dispatch
>>>>>>>>> > > [ 9%] Built target netcdf3
>>>>>>>>> > > [ 9%] Built target netcdf
>>>>>>>>> > > [ 10%] Built target ncgen
>>>>>>>>> > > [ 10%] Built target ncgen3
>>>>>>>>> > > [ 10%] Built target nccopy
>>>>>>>>> > > [ 10%] Built target ncdump
>>>>>>>>> > > [ 11%] Built target netcdff
>>>>>>>>> > > [ 11%] Built target xblas_build
>>>>>>>>> > > [ 12%] Built target blas
>>>>>>>>> > > [ 15%] Built target lapack
>>>>>>>>> > > [ 16%] Built target arpack
>>>>>>>>> > > [ 19%] Built target pnetcdf_fortran_obj
>>>>>>>>> > > [ 19%] Built target pnetcdf_c_obj
>>>>>>>>> > > [ 19%] Built target pnetcdf
>>>>>>>>> > > [ 19%] Built target ncvalid
>>>>>>>>> > > [ 19%] Built target boost_build
>>>>>>>>> > > [ 19%] Built target amber_common
>>>>>>>>> > > [ 20%] Built target rism
>>>>>>>>> > > [ 20%] Built target libpbsa
>>>>>>>>> > > [ 20%] Built target sff_rism_interface
>>>>>>>>> > > [ 20%] Built target sff_lex
>>>>>>>>> > > [ 20%] Built target dsarpack_obj
>>>>>>>>> > > [ 20%] Built target sff
>>>>>>>>> > > [ 21%] Built target gbnsr6
>>>>>>>>> > > [ 21%] Built target cifparse
>>>>>>>>> > > [ 22%] Built target addles
>>>>>>>>> > > [ 23%] Built target rism_mpi
>>>>>>>>> > > [ 23%] Built target sander_rism_interface
>>>>>>>>> > > [ 25%] Built target sqm_common
>>>>>>>>> > > [ 25%] Built target libsqm
>>>>>>>>> > > [ 26%] Built target sff_fortran
>>>>>>>>> > > [ 26%] Building Fortran object
>>>>>>>>> > >
>>>>>>>>> AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o
>>>>>>>>> > >
>>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1571.1:
>>>>>>>>> > > Included at
>>>>>>>>> > >
>>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>>> > >
>>>>>>>>> > > \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>>>> \xC2\xA0!
>>>>>>>>> > \xC2\xA0Fe
>>>>>>>>> > > rad
>>>>>>>>> > > 1
>>>>>>>>> > > Error: Invalid character in name at (1)
>>>>>>>>> > >
>>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread2.F90:1572.1:
>>>>>>>>> > > Included at
>>>>>>>>> > >
>>>>>>>>> /home/exx/Downloads/amber20_src/AmberTools/src/sander/mdread.F90:24:
>>>>>>>>> > >
>>>>>>>>> > > \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0 \xC2\xA0
>>>>>>>>> > x(L165-1+i)
>>>>>>>>> > > = 1
>>>>>>>>> > > 1
>>>>>>>>> > > Error: Invalid character in name at (1)
>>>>>>>>> > > make[2]: ***
>>>>>>>>> > >
>>>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/mdread.F90.o]
>>>>>>>>> > Error 1
>>>>>>>>> > > make[1]: ***
>>>>>>>>> [AmberTools/src/sander/CMakeFiles/sander_base_obj.dir/all]
>>>>>>>>> > > Error 2
>>>>>>>>> > > make: *** [all] Error 2
>>>>>>>>> > > [2]+ Done gedit run_cmake
>>>>>>>>> > >
>>>>>>>>> > > On Mon, Dec 14, 2020 at 8:36 PM Elvis Martis <
>>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>>> > > wrote:
>>>>>>>>> > >
>>>>>>>>> > > > One more thing I missed,
>>>>>>>>> > > > After you have successfully located the file and edited it
>>>>>>>>> accordingly,
>>>>>>>>> > > > please don't forget to recompile AMBER.
>>>>>>>>> > > > Best Regards
>>>>>>>>> > > > Elvis
>>>>>>>>> > > >
>>>>>>>>> > > >
>>>>>>>>> > > >
>>>>>>>>> > > > On Mon, 14 Dec 2020 at 17:05, Elvis Martis <
>>>>>>>>> elvis_bcp.elvismartis.in>
>>>>>>>>> > > > wrote:
>>>>>>>>> > > >
>>>>>>>>> > > > > OK.
>>>>>>>>> > > > > Have you compiled AMBER20 using the legacy configure
>>>>>>>>> method or the
>>>>>>>>> > new
>>>>>>>>> > > > > cmake method?
>>>>>>>>> > > > > If you have using the new cmake method of compilation,
>>>>>>>>> then I guess
>>>>>>>>> > you
>>>>>>>>> > > > > should find that file in
>>>>>>>>> > amber20_src/AmberTools/src/sander/mdread2.F90.
>>>>>>>>> > > > > if you have compiled using the legacy method, then you
>>>>>>>>> files should
>>>>>>>>> > in
>>>>>>>>> > > > the
>>>>>>>>> > > > > path where I sent previously.
>>>>>>>>> > > > > Best Regards
>>>>>>>>> > > > > Elvis
>>>>>>>>> > > > >
>>>>>>>>> > > > >
>>>>>>>>> > > > >
>>>>>>>>> > > > > On Mon, 14 Dec 2020 at 16:55, Vaibhav Dixit <
>>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>>> > >
>>>>>>>>> > > > > wrote:
>>>>>>>>> > > > >
>>>>>>>>> > > > >> Dear All and Elivs,
>>>>>>>>> > > > >> I'm afraid, I still can't find this file inside the
>>>>>>>>> AmberTools
>>>>>>>>> > folder,
>>>>>>>>> > > > >> there is no folder by the same sander.
>>>>>>>>> > > > >> All tests passed while installing Amber20, thus I think
>>>>>>>>> there is
>>>>>>>>> > some
>>>>>>>>> > > > >> confusion regarding the directory structure and file
>>>>>>>>> locations in
>>>>>>>>> > > > Amber18
>>>>>>>>> > > > >> and Amber20 as well.
>>>>>>>>> > > > >> Please let me know if you know the fix to this as well.
>>>>>>>>> > > > >> thank you and best regards.
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> (base) [exx.c107739 amber20]$ ls
>>>>>>>>> > > > >> $AMBERHOME/AmberTools/src/sander/mdread2.F90
>>>>>>>>> > > > >> ls: cannot access
>>>>>>>>> > > > >>
>>>>>>>>> /home/exx/Downloads/amber20/AmberTools/src/sander/mdread2.F90: No
>>>>>>>>> > such
>>>>>>>>> > > > >> file
>>>>>>>>> > > > >> or directory
>>>>>>>>> > > > >> (base) [exx.c107739 amber20]$ cd AmberTools/src/
>>>>>>>>> > > > >> (base) [exx.c107739 src]$ ls -arlth
>>>>>>>>> > > > >> total 28K
>>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 cpptraj
>>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 mm_pbsa
>>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 FEW
>>>>>>>>> > > > >> drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 pytraj
>>>>>>>>> > > > >> drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 ..
>>>>>>>>> > > > >> -rw-r--r--. 1 exx exx 1.9K Dec 4 17:26 config.h
>>>>>>>>> > > > >> drwxrwxr-x. 6 exx exx 4.0K Dec 4 17:45 .
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> On Mon, Dec 14, 2020 at 7:46 PM Elvis Martis <
>>>>>>>>> > > elvis_bcp.elvismartis.in>
>>>>>>>>> > > > >> wrote:
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> > Hi
>>>>>>>>> > > > >> > Sorry my bad. For AMBER20
>>>>>>>>> > > > >> > you can look in
>>>>>>>>> > > > >> > $AMBERHOME/AmberTools/src/sander/mdread2.F90
>>>>>>>>> > > > >> > Start looking from line number 1508 (this should be
>>>>>>>>> same unless
>>>>>>>>> > > there
>>>>>>>>> > > > >> were
>>>>>>>>> > > > >> > changes made earlier)
>>>>>>>>> > > > >> > this line starts with " else if ( gbsa == 2 ) then"
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >> > Best Regards
>>>>>>>>> > > > >> > Elvis
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >> > On Mon, 14 Dec 2020 at 15:57, Vaibhav Dixit <
>>>>>>>>> > > vaibhavadixit.gmail.com>
>>>>>>>>> > > > >> > wrote:
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >> > > Dear All,
>>>>>>>>> > > > >> > > I got this suggestion earlier last month also.
>>>>>>>>> > > > >> > > But to my surprise, I can't find these specified
>>>>>>>>> folders and
>>>>>>>>> > files
>>>>>>>>> > > > in
>>>>>>>>> > > > >> the
>>>>>>>>> > > > >> > > expected locations.
>>>>>>>>> > > > >> > > I ran a find command to search for the mdread.f file,
>>>>>>>>> but it is
>>>>>>>>> > > not
>>>>>>>>> > > > >> found
>>>>>>>>> > > > >> > > anywhere inside amber20 folder and subfolders.
>>>>>>>>> > > > >> > > This command works and finds other files as shown
>>>>>>>>> below.
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > Please do let me know what I'm missing to understand
>>>>>>>>> here.
>>>>>>>>> > > > >> > > thanks
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ ls -arlth
>>>>>>>>> > > > >> > > total 128K
>>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 7.5K Dec 6 2018 GNU_LGPL_v3
>>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 116 Dec 6 2018
>>>>>>>>> amber-interactive.sh
>>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 1.1K Apr 28 2020 Makefile
>>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 37K Apr 28 2020 LICENSE
>>>>>>>>> > > > >> > > drwxrwxr-x. 8 exx exx 4.0K Dec 4 15:17 licenses
>>>>>>>>> > > > >> > > drwxrwxr-x. 2 exx exx 4.0K Dec 4 15:17 doc
>>>>>>>>> > > > >> > > drwxrwxr-x. 19 exx exx 4.0K Dec 4 15:17 miniconda
>>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 share
>>>>>>>>> > > > >> > > drwxrwxr-x. 9 exx exx 4.0K Dec 4 15:17 benchmarks
>>>>>>>>> > > > >> > > drwxrwxr-x. 16 exx exx 4.0K Dec 4 15:17 dat
>>>>>>>>> > > > >> > > drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 AmberTools
>>>>>>>>> > > > >> > > -rw-r--r--. 1 exx exx 1.9K Dec 4 17:26 config.h
>>>>>>>>> > > > >> > > drwxrwxr-x. 8 exx exx 4.0K Dec 10 15:54 logs
>>>>>>>>> > > > >> > > drwxrwxr-x. 157 exx exx 4.0K Dec 10 15:58 test
>>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 1.8K Dec 10 20:38 amber.sh
>>>>>>>>> > > > >> > > -rwxr-xr-x. 1 exx exx 1.9K Dec 10 20:38 amber.csh
>>>>>>>>> > > > >> > > drwxrwxr-x. 15 exx exx 4.0K Dec 10 21:50 .
>>>>>>>>> > > > >> > > drwxr-xr-x. 2 exx exx 4.0K Dec 10 21:50 include
>>>>>>>>> > > > >> > > drwxrwxr-x. 4 exx exx 4.0K Dec 10 21:50 lib64
>>>>>>>>> > > > >> > > drwxr-xr-x. 4 exx exx 4.0K Dec 10 21:50 lib
>>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 10 21:50 bin
>>>>>>>>> > > > >> > > drwxr-xr-x. 24 exx exx 4.0K Dec 12 12:13 ..
>>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ cd AmberTools/
>>>>>>>>> > > > >> > > (base) [exx.c107739 AmberTools]$ ls -arlth
>>>>>>>>> > > > >> > > total 28K
>>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:17 benchmarks
>>>>>>>>> > > > >> > > drwxrwxr-x. 6 exx exx 4.0K Dec 4 15:17 examples
>>>>>>>>> > > > >> > > drwxrwxr-x. 3 exx exx 4.0K Dec 4 15:25 .pytest_cache
>>>>>>>>> > > > >> > > drwxrwxr-x. 7 exx exx 4.0K Dec 4 15:25 .
>>>>>>>>> > > > >> > > drwxrwxr-x. 6 exx exx 4.0K Dec 4 17:45 src
>>>>>>>>> > > > >> > > drwxrwxr-x. 15 exx exx 4.0K Dec 10 21:50 ..
>>>>>>>>> > > > >> > > drwxrwxr-x. 65 exx exx 4.0K Dec 10 21:59 test
>>>>>>>>> > > > >> > > (base) [exx.c107739 AmberTools]$ cd ..
>>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name mdread.f
>>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name amber.sh
>>>>>>>>> > > > >> > > ./amber.sh
>>>>>>>>> > > > >> > > (base) [exx.c107739 amber20]$ find . -name am1bcc
>>>>>>>>> > > > >> > > ./bin/am1bcc
>>>>>>>>> > > > >> > > ./bin/wrapped_progs/am1bcc
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > On Mon, Dec 14, 2020 at 7:14 PM Elvis Martis <
>>>>>>>>> > > > >> elvis_bcp.elvismartis.in>
>>>>>>>>> > > > >> > > wrote:
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > > Hi
>>>>>>>>> > > > >> > > > Here is a solution in a different thread, you will
>>>>>>>>> have to
>>>>>>>>> > edit
>>>>>>>>> > > > the
>>>>>>>>> > > > >> > > > mdread.f file. Please have a look at link below to
>>>>>>>>> know how to
>>>>>>>>> > > > edit
>>>>>>>>> > > > >> the
>>>>>>>>> > > > >> > > > file and where to find it.
>>>>>>>>> > > > >> > > > Re: [AMBER] bad atom type: Br from Jason Swails on
>>>>>>>>> 2011-11-11
>>>>>>>>> > > > (Amber
>>>>>>>>> > > > >> > > > Archive Nov 2011) (ambermd.org)
>>>>>>>>> > > > >> > > > <http://archive.ambermd.org/201111/0370.html>
>>>>>>>>> > > > >> > > > Best Regards
>>>>>>>>> > > > >> > > > Elvis
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > > > On Mon, 14 Dec 2020 at 15:34, Vaibhav Dixit <
>>>>>>>>> > > > >> vaibhavadixit.gmail.com>
>>>>>>>>> > > > >> > > > wrote:
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > > > > Dear All,
>>>>>>>>> > > > >> > > > > I'm getting the following error message while
>>>>>>>>> trying to run
>>>>>>>>> > > > >> MMPBSA.py
>>>>>>>>> > > > >> > > in
>>>>>>>>> > > > >> > > > > Amber20.
>>>>>>>>> > > > >> > > > > It says that the receptor mask overwritten with
>>>>>>>>> default,
>>>>>>>>> > > which I
>>>>>>>>> > > > >> > don't
>>>>>>>>> > > > >> > > > know
>>>>>>>>> > > > >> > > > > why it is doing even after I gave a specific
>>>>>>>>> receptor mask
>>>>>>>>> > > (1st
>>>>>>>>> > > > >> > residue
>>>>>>>>> > > > >> > > > is
>>>>>>>>> > > > >> > > > > my ligand in the trajectory). It also gives the
>>>>>>>>> bad atom
>>>>>>>>> > type
>>>>>>>>> > > > >> error,
>>>>>>>>> > > > >> > > > that
>>>>>>>>> > > > >> > > > > I got with Amber18 last month.
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > >From the output files (attached), it is not
>>>>>>>>> clear what is
>>>>>>>>> > the
>>>>>>>>> > > > >> exact
>>>>>>>>> > > > >> > > > nature
>>>>>>>>> > > > >> > > > > of the error, thus I have no clue how to proceed
>>>>>>>>> to fix the
>>>>>>>>> > > > same.
>>>>>>>>> > > > >> > > > > Can you please help me understand why the dry
>>>>>>>>> prmtop is not
>>>>>>>>> > > > >> > compatible?
>>>>>>>>> > > > >> > > > > And how can I possibly attempt to get this
>>>>>>>>> analysis done?
>>>>>>>>> > > > >> > > > > My mmpbsa input file is given below.
>>>>>>>>> > > > >> > > > > Thanking you all in advance.
>>>>>>>>> > > > >> > > > > Best regards
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > mmpbsa input file:
>>>>>>>>> > > > >> > > > > (base) [exx.c107739 mmpbsa-6701-tyr]$ more
>>>>>>>>> mmpbsa-decomp.in
>>>>>>>>> > > > >> > > > > &general
>>>>>>>>> > > > >> > > > > startframe=1, endframe=100, interval=5,
>>>>>>>>> > > > >> > > > > verbose=2, keep_files=2, netcdf=1,
>>>>>>>>> receptor_mask=2-386,
>>>>>>>>> > > > >> > > > > strip_mask=:387-16750,
>>>>>>>>> > > > >> > > > > /
>>>>>>>>> > > > >> > > > > &gb
>>>>>>>>> > > > >> > > > > igb=5, saltcon=0.150,
>>>>>>>>> > > > >> > > > > /
>>>>>>>>> > > > >> > > > > &pb
>>>>>>>>> > > > >> > > > > istrng=0.15, fillratio=4.0,
>>>>>>>>> > > > >> > > > > /
>>>>>>>>> > > > >> > > > > &decomp
>>>>>>>>> > > > >> > > > > idecomp=1, print_res="56-57; 74-75; 167; 222;
>>>>>>>>> 225-227;
>>>>>>>>> > > > 229-231;
>>>>>>>>> > > > >> > > > 272-273;
>>>>>>>>> > > > >> > > > > 275-277; 336; 368; 370; 372-373"
>>>>>>>>> > > > >> > > > > dec_verbose=1,
>>>>>>>>> > > > >> > > > > /
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Error message on the terminal
>>>>>>>>> > > > >> > > > > (base) [exx.c107739 mmpbsa-6701-tyr]$ MMPBSA.py
>>>>>>>>> -O -i
>>>>>>>>> > > > >> > mmpbsa-decomp.in
>>>>>>>>> > > > >> > > > -o
>>>>>>>>> > > > >> > > > > mmpbs-rouf-6701-tyr.dat -do
>>>>>>>>> decomp-rouf-6701-tyr.dat -sp
>>>>>>>>> > > > >> > > > > ../rouf-6701-tyr-solv.prmtop -cp
>>>>>>>>> ../rouf-6701-tyr-dry.prmtop
>>>>>>>>> > > -lp
>>>>>>>>> > > > >> > > > > ../tyr-dry.prmtop -y ../
>>>>>>>>> rouf-6701-tyr-solv-prod300.nc -rp
>>>>>>>>> > > > >> > > > > ../rouf-6701-nolig-dry.prmtop
>>>>>>>>> > > > >> > > > > Loading and checking parameter files for
>>>>>>>>> compatibility...
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >>
>>>>>>>>> > > >
>>>>>>>>> > >
>>>>>>>>> >
>>>>>>>>> */home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py:603:
>>>>>>>>> > > > >> > > > > UserWarning: receptor_mask overwritten with
>>>>>>>>> default*
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > warnings.warn('receptor_mask overwritten with
>>>>>>>>> default\n')
>>>>>>>>> > > > >> > > > > cpptraj found! Using
>>>>>>>>> /home/exx/Downloads/amber20/bin/cpptraj
>>>>>>>>> > > > >> > > > > sander found! Using
>>>>>>>>> /home/exx/Downloads/amber20/bin/sander
>>>>>>>>> > > > >> > > > > Preparing trajectories for simulation...
>>>>>>>>> > > > >> > > > > 20 frames were processed by cpptraj for use in
>>>>>>>>> calculation.
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Running calculations on normal system...
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Beginning GB calculations with
>>>>>>>>> > > > >> /home/exx/Downloads/amber20/bin/sander
>>>>>>>>> > > > >> > > > > calculating complex contribution...
>>>>>>>>> > > > >> > > > > * bad atom type: M1*
>>>>>>>>> > > > >> > > > > File
>>>>>>>>> "/home/exx/Downloads/amber20/bin/MMPBSA.py", line
>>>>>>>>> > 100,
>>>>>>>>> > > in
>>>>>>>>> > > > >> > > <module>
>>>>>>>>> > > > >> > > > > app.run_mmpbsa()
>>>>>>>>> > > > >> > > > > File
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >>
>>>>>>>>> > > >
>>>>>>>>> > >
>>>>>>>>> >
>>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/main.py",
>>>>>>>>> > > > >> > > > > line 218, in run_mmpbsa
>>>>>>>>> > > > >> > > > > self.calc_list.run(rank, self.stdout)
>>>>>>>>> > > > >> > > > > File
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >>
>>>>>>>>> > > >
>>>>>>>>> > >
>>>>>>>>> >
>>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>>>>>>>>> > > > >> > > > > line 82, in run
>>>>>>>>> > > > >> > > > > calc.run(rank, stdout=stdout, stderr=stderr)
>>>>>>>>> > > > >> > > > > File
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >>
>>>>>>>>> > > >
>>>>>>>>> > >
>>>>>>>>> >
>>>>>>>>> "/home/exx/Downloads/amber20/lib/python3.8/site-packages/MMPBSA_mods/calculation.py",
>>>>>>>>> > > > >> > > > > line 156, in run
>>>>>>>>> > > > >> > > > > raise CalcError('%s failed with prmtop %s!' %
>>>>>>>>> > > (self.program,
>>>>>>>>> > > > >> > > > > CalcError: /home/exx/Downloads/amber20/bin/sander
>>>>>>>>> failed
>>>>>>>>> > with
>>>>>>>>> > > > >> prmtop
>>>>>>>>> > > > >> > > > > ../rouf-6701-tyr-dry.prmtop!
>>>>>>>>> > > > >> > > > > Exiting. All files have been retained.
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > --
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Regards,
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Dr. Vaibhav A. Dixit,
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > Visiting Scientist at the Manchester Institute of
>>>>>>>>> > > Biotechnology
>>>>>>>>> > > > >> > (MIB),
>>>>>>>>> > > > >> > > > The
>>>>>>>>> > > > >> > > > > University of Manchester, 131 Princess Street,
>>>>>>>>> Manchester M1
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> > > > 7DN,
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> > > > >> UK
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> .
>>>>>>>>> > > > >> > > > > AND
>>>>>>>>> > > > >> > > > > Assistant Professor,
>>>>>>>>> > > > >> > > > > Department of Pharmacy,
>>>>>>>>> > > > >> > > > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>>> > > > >> > > > > Birla Institute of Technology and Sciences Pilani
>>>>>>>>> > > (BITS-Pilani),
>>>>>>>>> > > > >> > > > > VidyaVihar Campus, street number 41, Pilani,
>>>>>>>>> Rajasthan
>>>>>>>>> > 333031.
>>>>>>>>> > > > >> > > > > India.
>>>>>>>>> > > > >> > > > > Phone No. +91 1596 255652, Mob. No.
>>>>>>>>> +91-7709129400,
>>>>>>>>> > > > >> > > > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>>> > > > >> > vaibhavadixit.gmail.com
>>>>>>>>> > > > >> > > > >
>>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>>> > > > >> > > > >
>>>>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >>
>>>>>>>>> >
>>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > > P Please consider the environment before printing
>>>>>>>>> this
>>>>>>>>> > > > >> > > > > _______________________________________________
>>>>>>>>> > > > >> > > > > AMBER mailing list
>>>>>>>>> > > > >> > > > > AMBER.ambermd.org
>>>>>>>>> > > > >> > > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > > >> > > > >
>>>>>>>>> > > > >> > > > _______________________________________________
>>>>>>>>> > > > >> > > > AMBER mailing list
>>>>>>>>> > > > >> > > > AMBER.ambermd.org
>>>>>>>>> > > > >> > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > > >> > > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > --
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > Regards,
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > Dr. Vaibhav A. Dixit,
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > Visiting Scientist at the Manchester Institute of
>>>>>>>>> Biotechnology
>>>>>>>>> > > > (MIB),
>>>>>>>>> > > > >> > The
>>>>>>>>> > > > >> > > University of Manchester, 131 Princess Street,
>>>>>>>>> Manchester M1
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> > 7DN,
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> > > > UK
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+%0D%0A+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> .
>>>>>>>>> > > > >> > > AND
>>>>>>>>> > > > >> > > Assistant Professor,
>>>>>>>>> > > > >> > > Department of Pharmacy,
>>>>>>>>> > > > >> > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>>> > > > >> > > Birla Institute of Technology and Sciences Pilani
>>>>>>>>> (BITS-Pilani),
>>>>>>>>> > > > >> > > VidyaVihar Campus, street number 41, Pilani,
>>>>>>>>> Rajasthan 333031.
>>>>>>>>> > > > >> > > India.
>>>>>>>>> > > > >> > > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>>> > > > >> > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>>> > > > >> vaibhavadixit.gmail.com
>>>>>>>>> > > > >> > >
>>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>>> > > > >> > > https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > >
>>>>>>>>> > > >
>>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > > P Please consider the environment before printing
>>>>>>>>> this e-mail
>>>>>>>>> > > > >> > > _______________________________________________
>>>>>>>>> > > > >> > > AMBER mailing list
>>>>>>>>> > > > >> > > AMBER.ambermd.org
>>>>>>>>> > > > >> > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > > >> > >
>>>>>>>>> > > > >> > _______________________________________________
>>>>>>>>> > > > >> > AMBER mailing list
>>>>>>>>> > > > >> > AMBER.ambermd.org
>>>>>>>>> > > > >> > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > > >> >
>>>>>>>>> > > > >>
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> --
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> Regards,
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> Dr. Vaibhav A. Dixit,
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> Visiting Scientist at the Manchester Institute of
>>>>>>>>> Biotechnology
>>>>>>>>> > (MIB),
>>>>>>>>> > > > The
>>>>>>>>> > > > >> University of Manchester, 131 Princess Street,
>>>>>>>>> Manchester M1 7DN,
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> > UK
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+%0D%0A+UK?entry=gmail&source=g>
>>>>>>>>> .
>>>>>>>>> > > > >> AND
>>>>>>>>> > > > >> Assistant Professor,
>>>>>>>>> > > > >> Department of Pharmacy,
>>>>>>>>> > > > >> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>>> > > > >> Birla Institute of Technology and Sciences Pilani
>>>>>>>>> (BITS-Pilani),
>>>>>>>>> > > > >> VidyaVihar Campus, street number 41, Pilani, Rajasthan
>>>>>>>>> 333031.
>>>>>>>>> > > > >> India.
>>>>>>>>> > > > >> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>>> > > > >> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>>> > > vaibhavadixit.gmail.com
>>>>>>>>> > > > >> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>>> > > > >> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>> > > > >>
>>>>>>>>> > > > >>
>>>>>>>>> >
>>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>> > > > >>
>>>>>>>>> > > > >> P Please consider the environment before printing this
>>>>>>>>> > > > >> _______________________________________________
>>>>>>>>> > > > >> AMBER mailing list
>>>>>>>>> > > > >> AMBER.ambermd.org
>>>>>>>>> > > > >> http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > > >>
>>>>>>>>> > > > >
>>>>>>>>> > > > _______________________________________________
>>>>>>>>> > > > AMBER mailing list
>>>>>>>>> > > > AMBER.ambermd.org
>>>>>>>>> > > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > > >
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> > > --
>>>>>>>>> > >
>>>>>>>>> > > Regards,
>>>>>>>>> > >
>>>>>>>>> > > Dr. Vaibhav A. Dixit,
>>>>>>>>> > >
>>>>>>>>> > > Visiting Scientist at the Manchester Institute of
>>>>>>>>> Biotechnology (MIB),
>>>>>>>>> > The
>>>>>>>>> > > University of Manchester, 131 Princess Street, Manchester M1
>>>>>>>>> 7DN, UK
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>>>>> .
>>>>>>>>> > > AND
>>>>>>>>> > > Assistant Professor,
>>>>>>>>> > > Department of Pharmacy,
>>>>>>>>> > > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>>> > > Birla Institute of Technology and Sciences Pilani
>>>>>>>>> (BITS-Pilani),
>>>>>>>>> > > VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>>>>> > > India.
>>>>>>>>> > > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>>> > > Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>>> > > http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>>> > > https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>> > >
>>>>>>>>> > > ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>> > >
>>>>>>>>> > >
>>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>> > >
>>>>>>>>> > > P Please consider the environment before printing this e-mail
>>>>>>>>> > > _______________________________________________
>>>>>>>>> > > AMBER mailing list
>>>>>>>>> > > AMBER.ambermd.org
>>>>>>>>> > > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> > >
>>>>>>>>> > _______________________________________________
>>>>>>>>> > AMBER mailing list
>>>>>>>>> > AMBER.ambermd.org
>>>>>>>>> > http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>> >
>>>>>>>>>
>>>>>>>>>
>>>>>>>>> --
>>>>>>>>>
>>>>>>>>> Regards,
>>>>>>>>>
>>>>>>>>> Dr. Vaibhav A. Dixit,
>>>>>>>>>
>>>>>>>>> Visiting Scientist at the Manchester Institute of Biotechnology
>>>>>>>>> (MIB), The
>>>>>>>>> University of Manchester, 131 Princess Street, Manchester M1 7DN,
>>>>>>>>> UK
>>>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>>>>> .
>>>>>>>>> AND
>>>>>>>>> Assistant Professor,
>>>>>>>>> Department of Pharmacy,
>>>>>>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>>>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>>>>> India.
>>>>>>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>>>> vaibhavadixit.gmail.com
>>>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>>>
>>>>>>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>>>
>>>>>>>>>
>>>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>>>
>>>>>>>>> P Please consider the environment before printing this e-mail
>>>>>>>>> _______________________________________________
>>>>>>>>> AMBER mailing list
>>>>>>>>> AMBER.ambermd.org
>>>>>>>>> http://lists.ambermd.org/mailman/listinfo/amber
>>>>>>>>>
>>>>>>>>
>>>>>>>
>>>>>>> --
>>>>>>>
>>>>>>> Regards,
>>>>>>>
>>>>>>> Dr. Vaibhav A. Dixit,
>>>>>>>
>>>>>>> Visiting Scientist at the Manchester Institute of Biotechnology
>>>>>>> (MIB), The University of Manchester, 131 Princess Street,
>>>>>>> Manchester M1 7DN, UK
>>>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>>>> .
>>>>>>> AND
>>>>>>> Assistant Professor,
>>>>>>> Department of Pharmacy,
>>>>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>>>> India.
>>>>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
>>>>>>> vaibhavadixit.gmail.com
>>>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>>>
>>>>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>>>
>>>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>>>
>>>>>>> P Please consider the environment before printing this e-mail
>>>>>>>
>>>>>>>
>>>>>
>>>>> --
>>>>>
>>>>> Regards,
>>>>>
>>>>> Dr. Vaibhav A. Dixit,
>>>>>
>>>>> Visiting Scientist at the Manchester Institute of Biotechnology (MIB),
>>>>> The University of Manchester, 131 Princess Street, Manchester M1 7DN,
>>>>> UK
>>>>> <https://www.google.com/maps/search/131+Princess+Street,+Manchester+M1+7DN,+UK?entry=gmail&source=g>
>>>>> .
>>>>> AND
>>>>> Assistant Professor,
>>>>> Department of Pharmacy,
>>>>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>>>>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>>>>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>>>>> India.
>>>>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>>>>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
>>>>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>>>>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>>>>
>>>>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>>>>
>>>>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>>>>
>>>>> P Please consider the environment before printing this e-mail
>>>>>
>>>>>
>>>>
>>>> --
>>>> Best Regards
>>>> Elvis
>>>>
>>>
>>
>> --
>>
>> Regards,
>>
>> Dr. Vaibhav A. Dixit,
>>
>> Visiting Scientist at the Manchester Institute of Biotechnology (MIB),
>> The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
>> AND
>> Assistant Professor,
>> Department of Pharmacy,
>> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
>> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
>> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
>> India.
>> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
>> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
>> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
>> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>>
>> ORCID ID: https://orcid.org/0000-0003-4015-2941
>>
>> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>>
>> P Please consider the environment before printing this e-mail
>>
>>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: 02-image.png)
