Date: Fri, 26 Jun 2020 11:48:47 -0700
I have a total of 500 ns trajectory - 250 + 250 ns - starting from the same
eq.rst file. I calculated the RMSF (wrt to the first frame as the reference)
for this 500 ns using this cpptraj script:
parm ../../prod/pd1/wt.prmtop
trajin ../../prod/pd1/[1]wt_[2]pd.nc
trajin ../../prod/pd2/[3]wt_[4]pd.nc
reference ../../prod/pd1/[5]wt_[6]pd.nc 1
rms .CA reference
atomicfluct out RMSF.dat .CA byres
run
I would expect the RMSF calculated using this script to give the average
RMSF from the two production runs but it seems that the RMSF calculated
using the input above is not the true average of the two trajectories.
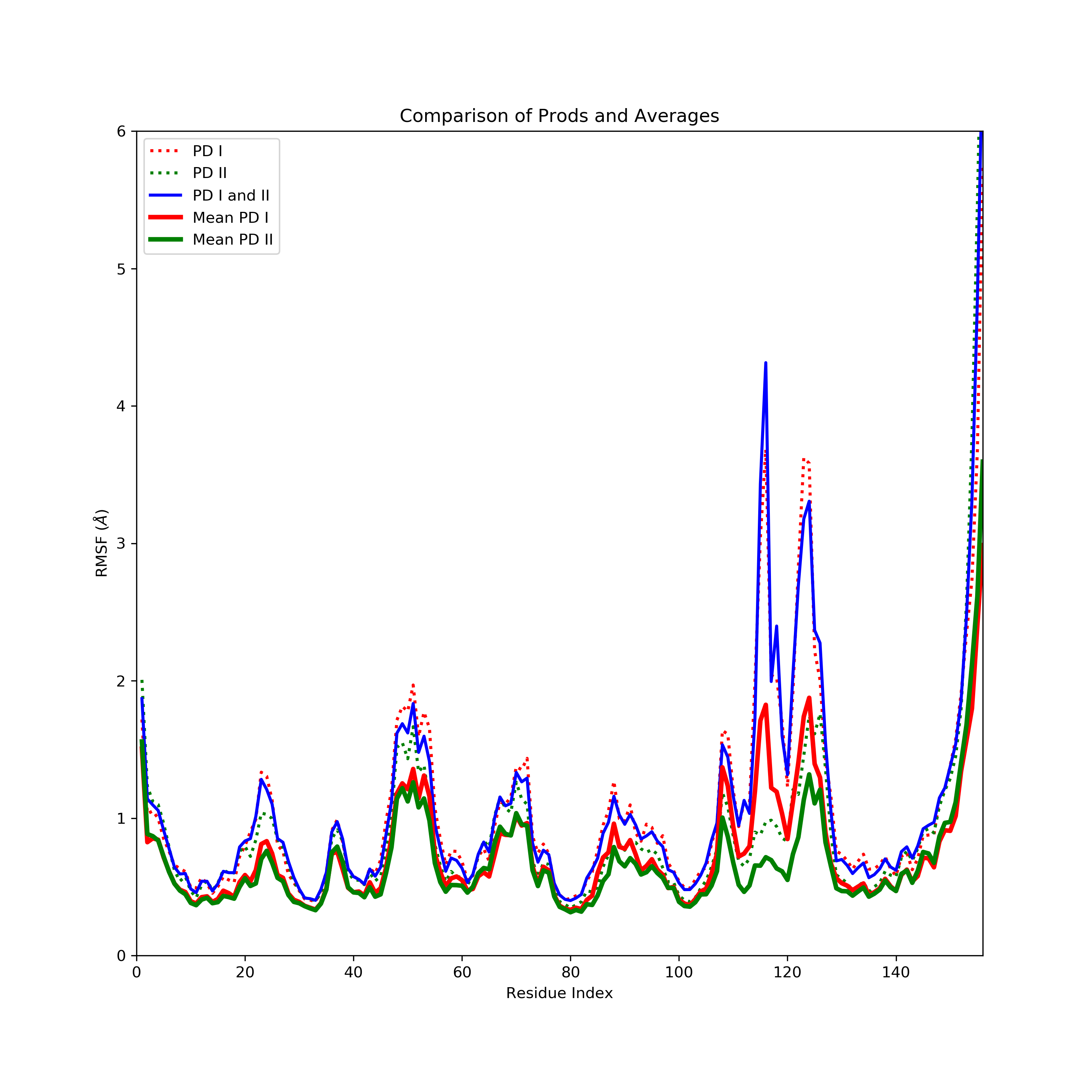
The attached graph will make it clearer.
comp_mean.png contains the RMSF calculated using cpptraj for the entire PD1
and PD2 and for PD1 + PD2. I have also plotted here the mean RMSFs (for 10ns
chunks) for PD 1 and PD 2 that I calculated using python. Am I using the
RMSF script incorrectly? I want to calculate the residue-wise fluctuations
of the Ca atoms of the peptide backbone of my entire simulation (i.e. PD 1
and 2 combined).
Thanks a lot in advance for all the help!
Kartik Lakshmi Rallapalli
References
1. https://urldefense.com/v3/__http:/wt_pd.nc__;!!Mih3wA!RkWVHZ9uzYaTsfRwSkKOl8GwVSF_aGB56sRRlI2gEzyf_i_Lj7VJPgwZCZ9m$
2. http://pd.nc/
3. https://urldefense.com/v3/__http:/wt_pd.nc__;!!Mih3wA!RkWVHZ9uzYaTsfRwSkKOl8GwVSF_aGB56sRRlI2gEzyf_i_Lj7VJPgwZCZ9m$
4. http://pd.nc/
5. https://urldefense.com/v3/__http:/wt_pd.nc__;!!Mih3wA!RkWVHZ9uzYaTsfRwSkKOl8GwVSF_aGB56sRRlI2gEzyf_i_Lj7VJPgwZCZ9m$
6. http://pd.nc/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: comp_mean.png)
