Date: Mon, 6 Jan 2020 09:32:52 +0530
Hiii David,
these are my tleap commands:
source leaprc.ff99SB
source leaprc.gaff
1bnr = sequence {NALA GLN VAL ILE ASN THR PHE ASP GLY VAL NALA ASP TYR LEU
GLN THR TYR HIS LYS LEU PRO ASP ASN TYR ILE THR LYS SER GLU NALA GLN NALA
LEU GLY TRP VAL NALA SER LYS GLY ASN LEU NALA ASP VAL NALA PRO GLY LYS SER
ILE GLY GLY ASP ILE PHE SER ASN ARG GLU GLY LYS LEU PRO GLY LYS SER GLY ARG
THR TRP ARG GLU NALA ASP ILE ASN TYR THR SER GLY PHE ARG ASN SER ASP ARG
ILE LEU TYR SER SER ASP TRP LEU ILE TYR LYS THR THR ASP HIS TYR GLN THR PHE
THR LYS ILE CARG}
saveoff 1bnr 1bnr_linear.lib
saveoff 1bnr 1bnr_linear.pdb
loadoff 1bnr 1bnr_linear.lib
saveamberparm 1bnr 1bnr_linear.prmtop 1bnr_linear.0.inpcrd
quit
~i intend to build a linear structure of these residues...
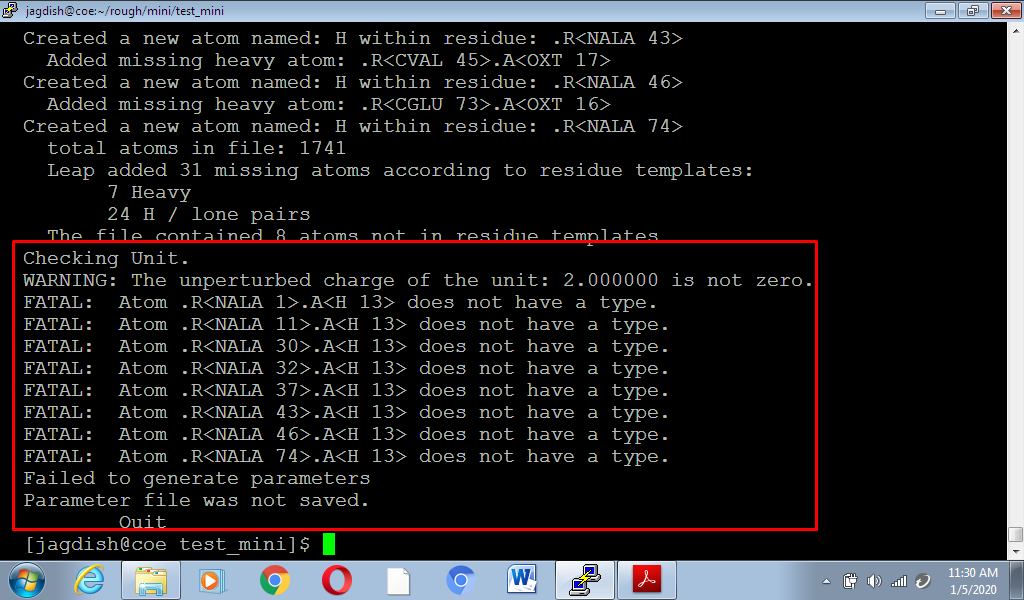
my output is :
attached below:
thanks
On Mon, Jan 6, 2020 at 12:03 AM David Case <david.case.rutgers.edu> wrote:
> On Sun, Jan 05, 2020, ankita mehta wrote:
>
> > what is the solution to remove TER inserted before ALA?
> >It cinsiders all alanine to be NALA?
>
> Just to repeat: we have no idea what sequence (or other commands) you
> used. It is generally far better to tell us what you actually did, not
> what your intent was. That is, don't say "I'm trying to build a pdb file
> through sequence command" (that describes your intent); rather say "here
> are the exact commands I gave to tleap, and here was the result." (that
> describes what you actually did.)
>
> In this particular case, as Carlos has noted, the structures Amber
> creates by default are not intended to be realisitic configurations.
>
> ....dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_6.png)
