Date: Thu, 16 Aug 2018 10:57:47 +0200
Hi all,
We are trying to implement the revised Tan, Piana, Dirks and Shaw RNA force
field in Amber (https://doi.org/10.1073/pnas.1713027115 ) .
I will briefly share with you our step, in order to receive some more
comments and insightful comments and also hopefully give a sort of overview
that could be useful to someone..
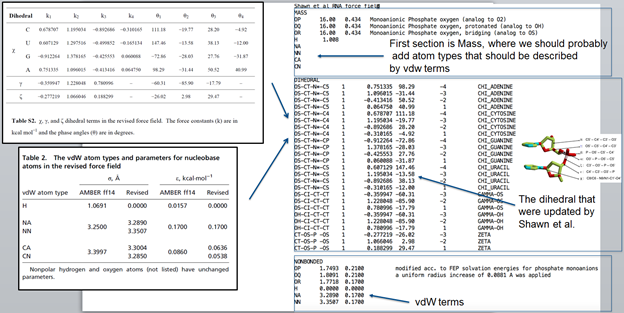
What we should do is add the modified electrostatic, vdW, and torsional
parameters in the AMBER ff14 RNA force field
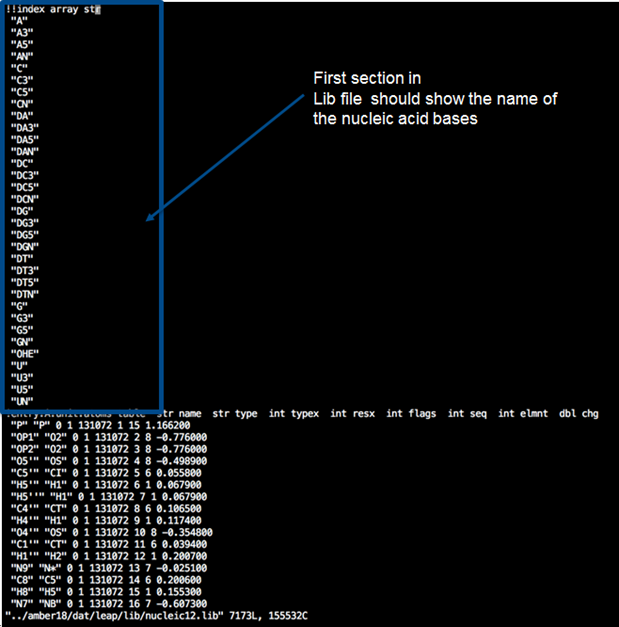
*LIB file:*
1. Copy the library file “nucleic12.lib” in
“/amber18/dat/leap/lib/nucleic12.lib” and rename it to RNA.lib
2. Delete all entries that include DNA (*i.e.* I think delete all the
one that starts with “D***”)
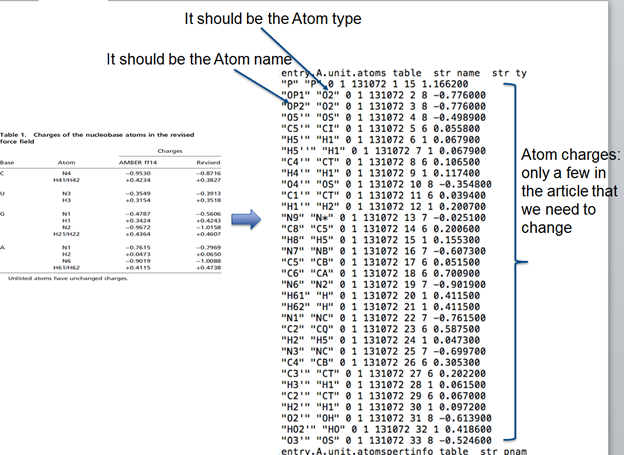
3. Modify the charges
*Just a couple of questions:*
*1. **Do the charges in the article table correspond to “Atom name”
column or “atom type” in the .lib column?*
*2. **Is the atom type between RNA and DNA redundant? If yes, do you
have any idea how to distinguish between RNA and DNA?*
*FRCMOD file:*
1. Add revised dihedral angle
2. Add revised vdW parameters
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: 02-image.png)
(image/png attachment: 03-image.png)
