Date: Thu, 29 Jun 2017 09:17:58 +0000
Dear Amber experts,
I have Amber16 and Amber Tools16.
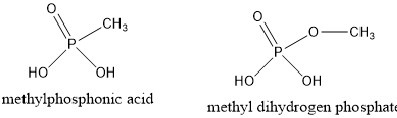
I need to simulate by constant pH MD complex of protein
with ligands containing phosphonic and phosphate
functional groups like structural examples presented in figure below.
Unfortunately I didnít find in amber documents and tutorials how to prepare
for constant pH MD input files containing such functional groups.
Your help highly appreciated.
Thank you,
Michael
[cid:5878e43b-68e1-447c-9aa4-eb0655daf19e]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: Picture1.jpg)
