Date: Thu, 17 Dec 2015 13:02:27 +0100
Hi Trang,
The first thing I notice about your files is that you appear to be using
protein atom types. This only matter if you modify parameters that are also
present in the protein force field. Generally, you should a) use the Gaff
atom types for ligands, or b) create new atom types that will prevent
overwriting the protein parameters. Option a) is easier.
(I couldn't understand your figure, since there were no atom labels and no
perspective to show depth. You also didn't indicate if the attached pep.pdb
file was the NMR structure or one from the MD simulation.)
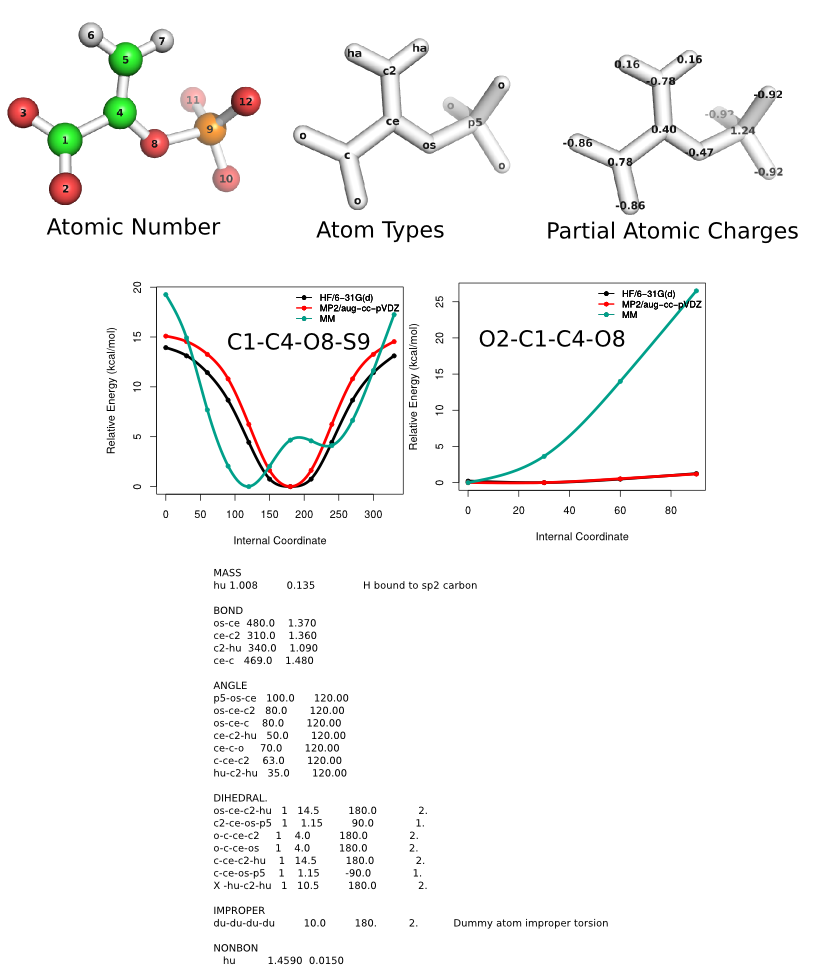
With that said, I went ahead and generated some QM target data and your
ligand with Gaff atom types. If I use your parameters (which I think I
converted correctly by matching your atom types with the Gaff atom types),
you would obtain the attached molecular mechanics (MM) curves. Your
parameters predict two C1-C4-O8-S9 minima. Based on the gas-phase QM data,
the molecule has one C1-C4-O8-S9 minma at a 180 degrees. The barrier
O2-C1-C4-O8 rotation is low, with an MP2 value of 1.2 kcal/mol. One could
to a scan of both dihedrals to generate a surface, but I don't believe a
second minima would appear.
The QM data was computed at HF/6-31G(d)//HF/6-31G(d) and
MP2/aug-cc-pVDZ//HF/6-31G(d) theory levels. The partial atomic charges are
RESP (2-stage 0.0005 and 0.001) fit to HF/6-31G(d) Connolly molecular
electrostatic potentials, using the HF/6-31G(d)//HF/6-31G(d) fully
optimized geometry. This charge set should be compatible with the Amber
protein residues.
Hope that helps.
On Wed, Dec 16, 2015 at 3:07 PM, Trang Nguyen <trangnt.iastate.edu> wrote:
> Hi Amber-developers/users,
>
> Our group currently works with Enzyme I-PTS (monomer, 313aa) and ligand PEP
> (12atoms, -3 charge). The original model EI-PEP comes from our both
> supposition and NMR experiments. After running MD simulation for 1ns, we
> recognized that the ligand's structure has been changed significantly from
> the original structure (see attached file). To our point of view, this
> change in ligand's structure should not be acceptable, compared with the
> experimental model. Especially in case the change is caused mainly by
> solving bad contacts during system preparation. Now, we are working on
> different reasons causing the problem.
>
> - Sets of charge on ligand. We tried with different charge methods to see
> which performs better during simulations. We tried to get RESP charges with
> GAMESS-US output but failed. We are not sure that the providing script (of
> Dr Hans De Winter) works for GAMESS-US or PC-GAMESS. We also would like to
> try with R.E.D. server at http://q4md-forcefieldtools.org/REDS/ but it
> requires name and password for authorization.
>
> - Angle/bond/dihedral definition. It seems that we have to define our own
> lib and frcmod files for ligand. Then the ligand's structure after 1ns
> looks better (not changed so much with the experimental model). But could
> you please help me to check these parameters (please see the attached files
> for pdb, lib as "log" and frcmod)
>
> - The final thing are parameters when running MD simulation from
> minimization to production. We did minimize system with strong restraint on
> both protein and ligand (at the first time, by following some online
> tutorials, we only restraint backbone of protein), then minimize with soft
> restraint and fully relax. For heating, density and equilibrium, we also
> tried to perform the same thing: strong restraint then relax gradually.
> Btw, can we perform restraint with different forces for different groups?
> i.e. strongly restraint on ligand, and weakly restraint on backbone.
>
> Could you please give me any suggestion. Do these approaches sound
> reasonable to get through the problem.
>
> Thank you very much
>
> Best Regards,
> Trang Nguyen
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
>
-- Karl. N. Kirschner, Ph.D. Research Associate Bonn-Rhein-Sieg University of Applied Sciences Grantham-Allee 20, 54757 Sankt Augustin, Germany
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: data.png)
