Date: Fri, 13 Nov 2015 17:17:37 +0530
Sorry, I forgot to attach the file.
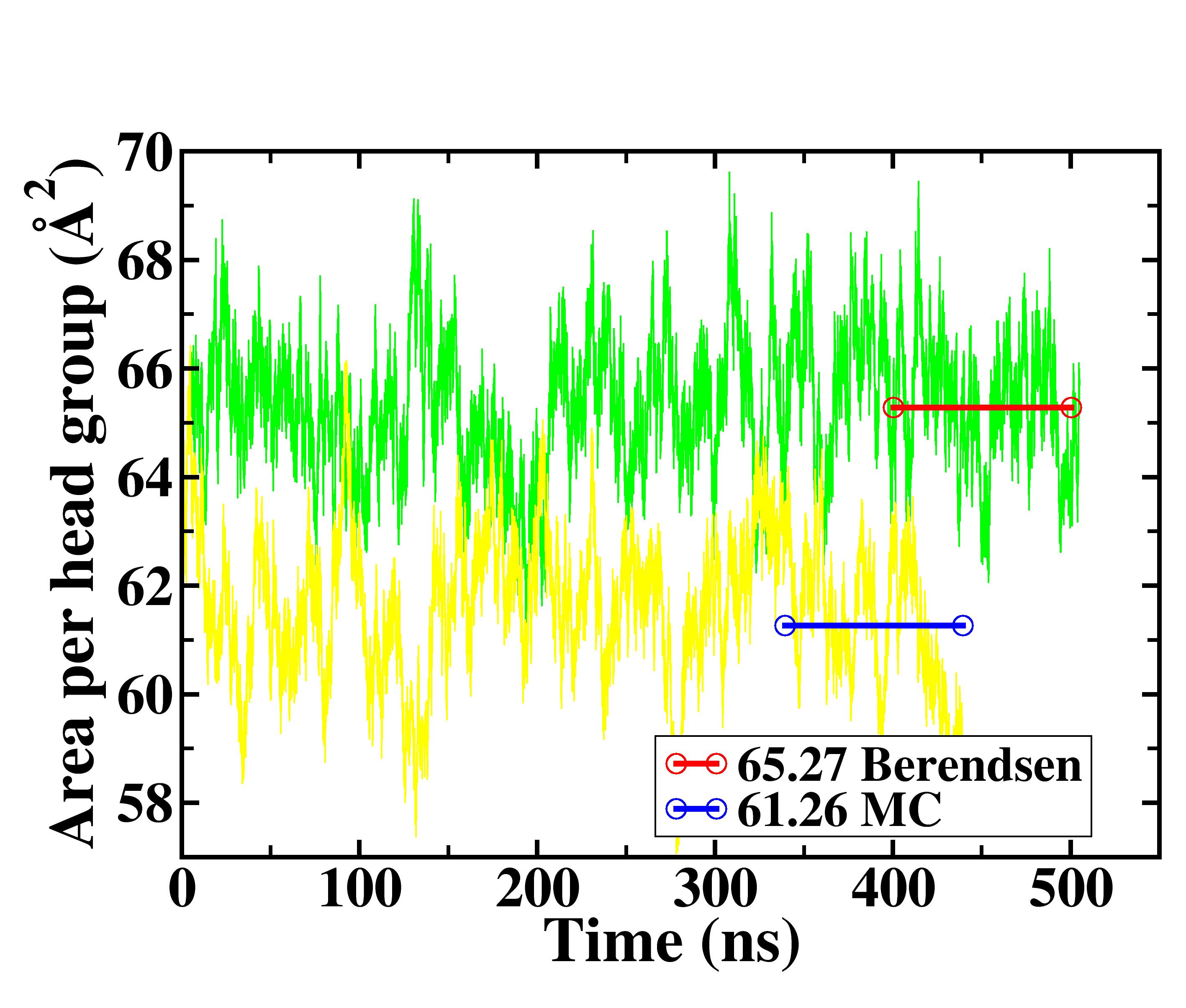
Here is the image showing the APH variation of
lipid with simulation time.
Thanks
On Fri, Nov 13, 2015 at 5:12 PM, Himanshu Joshi <himanshuphy87.gmail.com>
wrote:
> Dear Friends,
> While running POPC bilayer simulation in AMBER14 (LIPID14, pmemd.cuda),
> I found very different Area per head group (APH) using two
> different barostat. I ran two identical lipid (64 lipid per leaflet)
> system.
> I see very less APH for MC babrostat (61.2) as compared to Berendsen
> barostat(~65.3 A) during the course of simulation. The berendson barostat
> seems to reproduce the APH reported in Lipid14 force-field paper but not
> the MC barostat.
>
> Is it expected ? Please refer to the attached figure for the variation of
> APH with simulation time. Rest of the input parameters are exactly similar
> except the barostat for both the systems. I follow the procedure given in
> the tutorial by Benjamin Madej and Ross Walker on the official AMBER
> website to do the simulation.
>
> Another thing I observed (in lipid +complex system) that if I use NTR=1
> option (Rest input parameter similar to tutorial) to restrain the system
> during the equilibration, the simulations are not stable.
>
> Can you please comment on above issues.
> Thanks for the kind concern.
>
>
> --
>
>
>
> *With Regards,HIMANSHU JOSHI Graduate Scholar, Center for Condense Matter
> TheoryDepartment of Physics IISc.,Bangalore India 560012*
>
-- *With Regards,HIMANSHU JOSHI Graduate Scholar, Center for Condense Matter TheoryDepartment of Physics IISc.,Bangalore India 560012*
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: com_npt.jpg)
