Date: Tue, 2 Dec 2014 21:58:41 +0000
Hello,
I am working on trying to attach a organic linker to two single stranded DNA at separate ends to mimic the experimental ring-like structure of the proximity effect. The linker is attached to the DNA through a phosphate group at the 5' and 3' ends. The organic linker has two capped phosphate groups. I compared my system to the AMBER tutorial A1. I developed RESP charges for the linker. Developing the parameters for the linker was a bit of a hassle. Then I proceeded to run the linker through LEAP. This was successful after I established unique atom types, but this was done through copying and pasting from a .mol2 file to the linker pdb file to run in LEAP. How do you develop unique atom types for the molecule using AMBER if it does not already do it? I was successful in running the DNA (polyAT decamer) in LEAP. LEAP did create a H5T atom but that could easily be removed by a visualizer method. I deleted one side of the DNA (polyAT to ployA). Before I proceeded to put them together, I deleted the phosphate capped ends from the linker and added a phosphate group to the 5' end of the DNA. I used Chimera combine command to link the structure. Using Join models, I highlighted the two ends I wanted attached and linked them together, unfortunately this software did not allow me to construct the whole complex. When I tried to run half of the created structure ,the biggest problem I feel is AMBER did not see the fixed group (phosphate group) that puts two individual structures together.
>From all this being said, is there a way to attach DNA to a organic linker, have readable atom types, notice the attached groups, develop parameters, and run the whole structure though LEAP to start simulations?
I know I am not the first to want to attach DNA to a linker at two separate ends. I know there is someone who can guide me in the right direction.
See attached files to get a general idea of what I am trying to accomplish.
Thank you and have a great day!
Brittany Boykin
Graduate Student
Auburn University
Department of Chemistry and Biochemistry
e: bzb0031.tigermail.auburn.edu
c: (404)545.1036
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
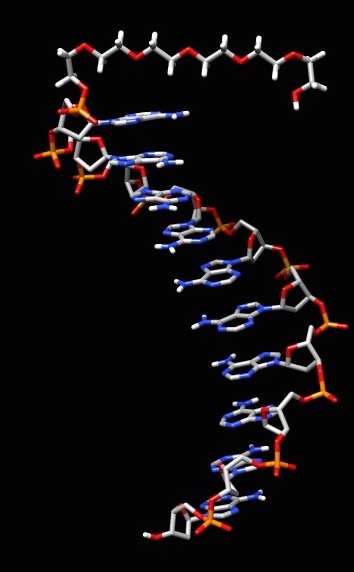
(image/jpeg attachment: DNA_linker_complex.jpg)
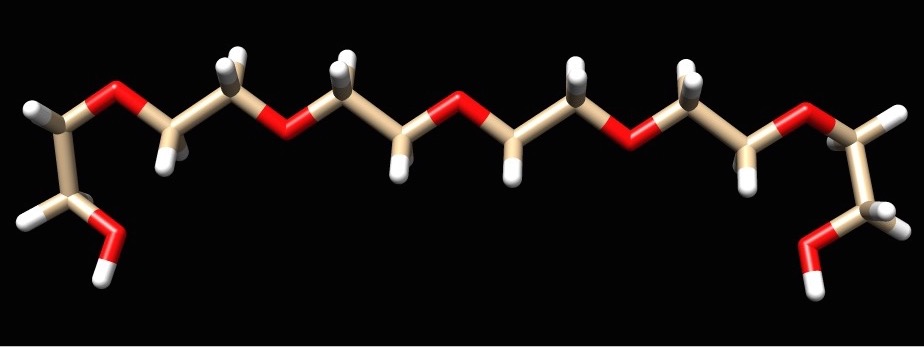
(image/jpeg attachment: linker-no-phos.jpg)
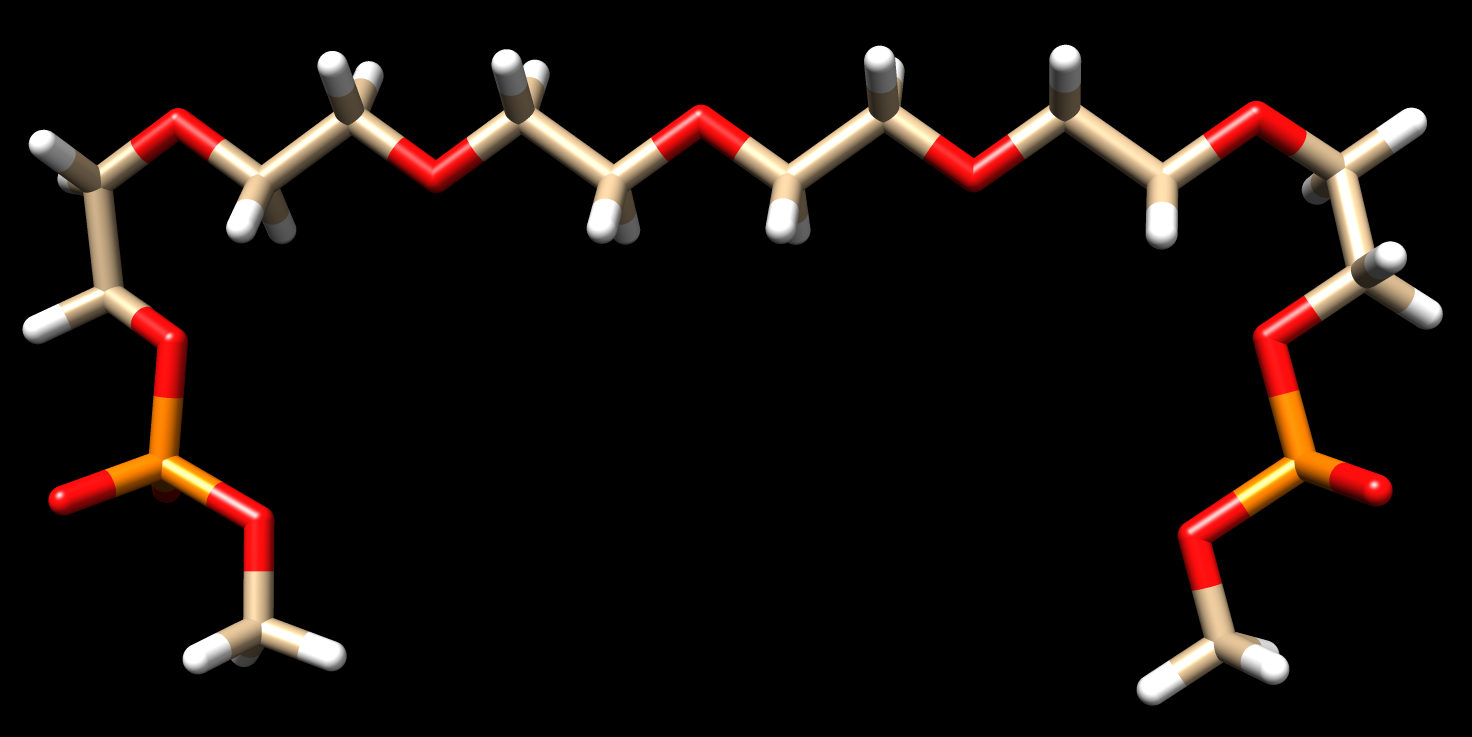
(image/png attachment: linker-with-phosphates.png)

(image/png attachment: polyA.png)
