Date: Tue, 12 Nov 2013 10:41:59 -0500
Hi Thomas,
1. I don't know how vmd is calculating those vectors (you could ask on
VMD mailing list), but note that angles of alpha, beta, gamma = 90 90
120 (what you have in your inpcrd) and 90 90 60 (what vmd molinfo
command gives) are compatible with each other, but this is not the same
as 60 90 90, which is the error on the webpage you reference (hopefully
soon to be corrected).
2. IFBOX=3 specifies a general triclinic system
3. I think part of the confusion lies in the fact that a hexagonal box,
an orthorombic box, etc are all special cases of a triclinic cell. So
simulating, for example, a box with angles 90 90 90 will yield the same
force/gradient calculations whether you tell your MD program that this
is a triclinic cell or you specify that it is orthorombic. As far as I
know, the only difference would be in simplifying some of the
calculations and how the system is imaged. The point is: if your box
vectors and angles are consistent with the system you want to have, d d
h 90 90 120 for a hexagonal space tasselation, then you are good.
4. To check if your system is set up correctly one thing you can do is
convert your inpcrd file to a pdb (use ambpdb) and add a CRYST1 pdb
format record. Then in vmd you can use the periodic tab in the
Graphics->Representations window to generate periodic boundy copies of
your system: if all space is neatly tasselated, you are good.
5. Also check density and energy during your simulation: if the box is
too small, for the coordinates you provide, periodic images will
generate steric clashes and energy will shoot up. If the box is too big,
you will have empty space in your system and density will drop down.
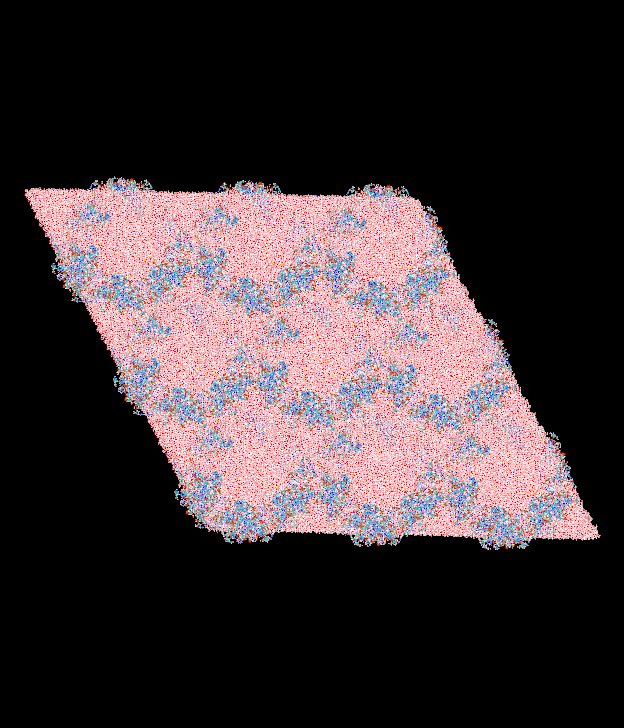
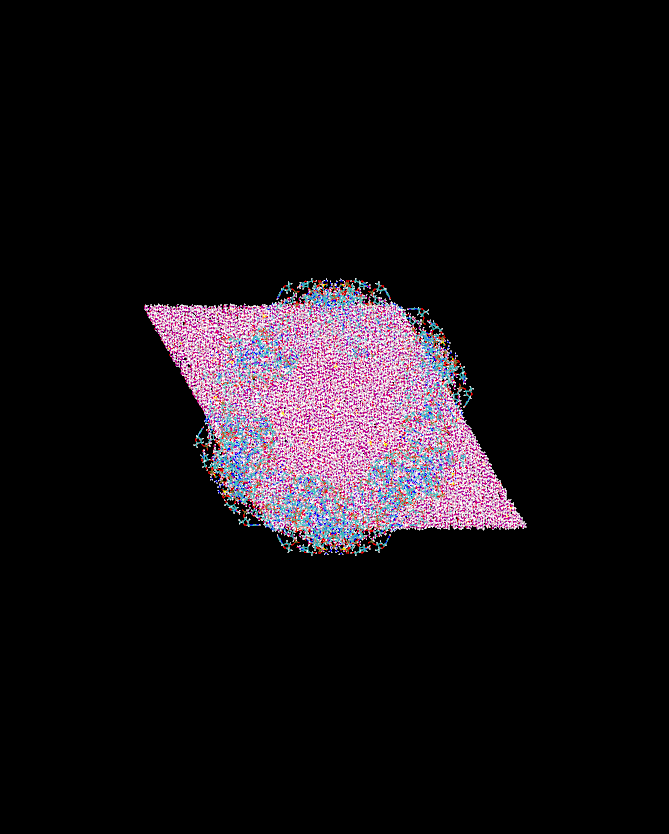
6.About your images: is that the same system? Why are there empty spaces
there between hexagonal shaped boxes? Again, the unit cell should look
like a triclinic cell and it is only the tasselation in space of those
unit cells that will create a hexagonal "look". (see attached images)
Pawel
On 11/12/2013 07:28 AM, Thomas Evangelidis wrote:
> I've missed a part of the previous message, see below:
>
>
> On 12 November 2013 14:11, Thomas Evangelidis <tevang3.gmail.com> wrote:
>
>> Brian & Pawel,
>>
>> Thanks for the information. I have done a simulation with a truncated
>> octahedron in the past in NAMD using AMBER .prmtop/.inpcrd, but that case
>> was easier due to solvateOct LeAP command.
>>
>> This time I create a protein-membrane-water system as a hexagona prism
>> with CHARMM-GUI, I modified the file as described in the tutorial and
>> passed it to LeAP. There I did:
>>
>>
> setBox complex centers
>
> ,but the box a,b,c vectors were overestimated, so I visually inspected the
> periodic cells in VMD and adjusted a,b,c to values given below. I also set
> IFBOX to 3 inside .prmtop. By the way, I know what 0,1 and 2 do, but what
> does IFBOX==3 mean?
>
>
>> Regarding the last line of .inpcrd, it seem that VMD perceives the lattice
>> vector angles differently. According to this post:
>>
>> http://archive.ambermd.org/200907/0035.html
>>
>> the last .inpcrd line correspond to "a b c alpha beta gamma". However, VMD
>> definition of alpha, beta and gamma is not exactly as in AMBER. See the
>> example below which shows the impact of NPT equilibration with NAMD were
>> the protein was constrained:
>>
>>
>> ## last .inpcrd line: 106.0000000 106.0000000 86.0000000 90.0000000
>> 90.0000000 120.000000
>> # box info at the beginning:
>> % molinfo top get {a b c}
>> 106.000000 106.000008 87.000000
>> % molinfo top get {alpha beta gamma}
>> 90.000000 90.000000 60.000004
>>
>> # box info at the end:
>> % molinfo top get {a b c}
>> 97.726463 98.022034 76.186539
>> % molinfo top get {alpha beta gamma}
>> 90.000000 90.000000 60.099697
>>
>> ## last .inpcrd line: 106.0000000 106.0000000 86.0000000 60.0000000
>> 90.0000000 90.000000
>> # box info at the beginning:
>> % molinfo top get {a b c}
>> 106.000000 106.000008 87.000000
>> % molinfo top get {alpha beta gamma}
>> 90.000000 90.000000 60.000004
>>
>> # box info at the end:
>> % molinfo top get {a b c}
>> 95.999474 95.565758 79.677513
>> % molinfo top get {alpha beta gamma}
>> 90.000000 90.000000 59.849758
>>
>>
>> As I can judge from the above results, "106.0000000 106.0000000
>> 86.0000000 90.0000000 90.0000000 120.000000" is equivalent to
>> "106.0000000 106.0000000 86.0000000 60.0000000 90.0000000 90.000000",
>> namely both Pawel and http://ambermd.org/namd/namd_amber.html are correct.
>>
>> I have uploaded snapshots of the system in the a-b plane before and after
> equilibration. It seems tome that the starting and final position are
> identical using both angle sets.
>
> https://www.dropbox.com/sh/i3f3y8xshbfkvy7/V7eSX2Cbr2
>
> Does anyone know the reason for the discrepancy in alpha,beta,gamma angles
> between VMD and AMBER (.inpcrd)?
> What variables should I monitor to see if I am simulating a correctly
> defined periodic system?
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: 9_hexagonal_boxes.png)
(image/png attachment: one_hexagonal_box.png)
