Date: Wed, 15 Nov 2023 18:35:47 -0800
I see. Indeed, the membrane probe is much larger than the water probe.
There is a mmpbsa_py test case for a realistic membrane protein-ligand
system. It's under
$AMBERHOME/AmberTools/test/mmpbsa_py/12_Membrane_Channel
In this test case, maxarcdot is set to be 10 times that of default value
(15,000) due to the much larger membrane probe.
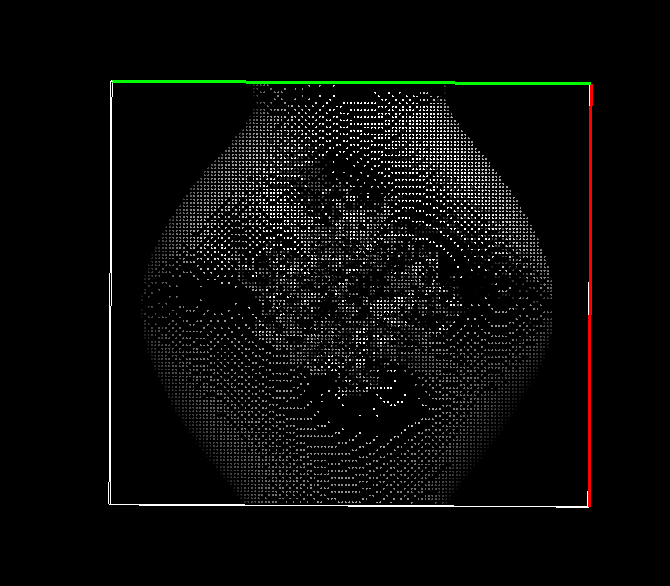
Also, your visualization doesn't seem to have a membrane. If you don't
mind, just email me your test files and input files off the list so I can
take a look.
All the best,
Ray
-- Ray Luo, Ph.D. Professor of Structural Biology/Biochemistry/Biophysics, Chemical and Materials Physics, Chemical and Biomolecular Engineering, Biomedical Engineering, and Materials Science and Engineering Department of Molecular Biology and Biochemistry University of California, Irvine, CA 92697-3900 On Wed, Nov 15, 2023 at 6:02 PM Matthew Guberman-Pfeffer < matthew.guberman-pfeffer.uconn.edu> wrote: > Hi Ray, > > That’s actually the part that surprised me: My system is a <150 residue > helix bundle protein with two heme groups. I only get the error related to > maxarcdot when trying to use an implicit membrane. Could that somehow be > related? > > I notice that the implicit membrane, when visualized by printing the level > set function, looks like a blob (see attached). Is this what it should look > like? The example int eh manual looks more rectangular. > > Thanks again for the help; I deeply appreciate it! > [image: Screenshot 2023-11-15 at 8.59.22 PM.png][image: Screenshot > 2023-11-15 at 8.59.56 PM.png] > Best, > Matthew > > > On Nov 15, 2023, at 8:52 PM, Ray Luo <rluo.uci.edu> wrote: > > *Message sent from a system outside of UConn.* > > > Matthew, > > I can't recommend anything out of the blue. You may add 250 each time > to see when it goes away. Apparently, the consequence of using a > larger number would drive up memory usage. There is no other effect on > the computation. > > You probably have a system whose packing is different from proteins. > It could also be due to the presence of many atoms with large radii. > > All the best, > Ray > -- > Ray Luo, Ph.D. > Professor of Structural Biology/Biochemistry/Biophysics, > Chemical and Materials Physics, Chemical and Biomolecular Engineering, > Biomedical Engineering, and Materials Science and Engineering > Department of Molecular Biology and Biochemistry > University of California, Irvine, CA 92697-3900 > > On Wed, Nov 15, 2023 at 4:12 PM Matthew Guberman-Pfeffer > <matthew.guberman-pfeffer.uconn.edu> wrote: > > > Ray, > > Thank you so much for the reply! What is a reasonable range of values? Is > there any harm in making maxarcdot too large? And, why would the number per > atom need to be larger. I’d like to understand a bit about what this > setting and issue means. Can you please help me understand this points. > > Best, > Matthew > > > On Nov 15, 2023, at 6:29 PM, Ray Luo via AMBER <amber.ambermd.org> wrote: > > *Message sent from a system outside of UConn.* > > > Matthew, > > This issue was fixed in the source code last time. You can now set the > keyword "maxarcdot" in the pb control name list. The default is 1500 > per atom. Looks like you'll have to set it to a larger number for your > system. > > All the best, > Ray > -- > Ray Luo, Ph.D. > Professor of Structural Biology/Biochemistry/Biophysics, > Chemical and Materials Physics, Chemical and Biomolecular Engineering, > Biomedical Engineering, and Materials Science and Engineering > Department of Molecular Biology and Biochemistry > University of California, Irvine, CA 92697-3900 > > On Wed, Nov 15, 2023 at 12:44 PM Matthew Guberman-Pfeffer via AMBER > <amber.ambermd.org> wrote: > > Dear Amber community, > > I get the the following error when trying to run PBSA with an implicit > membrane slab: > > SA Bomb in circle(): Stored surface points over limit****** > > A similar issue was reported nearly 20 years ago ( > https://nam10.safelinks.protection.outlook.com/?url=https%3A%2F%2Furldefense.com%2Fv3%2F__http%3A%2F%2Farchive.ambermd.org%2F200504%2F0365.html__%3B!!CzAuKJ42GuquVTTmVmPViYEvSg!M_sE739oXyJHj761ZXLacoC02jUOzCaTzaQ54kOXQHaOH8RCh9a8HiGWGvKWQm6Wj8lxHhMqMkY%24&data=05%7C01%7Cmatthew.guberman-pfeffer%40uconn.edu%7C3bc44cb73bf94ca3abde08dbe646ca54%7C17f1a87e2a254eaab9df9d439034b080%7C0%7C0%7C638356963936851278%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=XEPTZbK%2BH1Xmqi369JApzOB3YUS8vXzxY83dWlpHWfY%3D&reserved=0 > ), but a resolution was not given. Can any one please help me resolve > this problem? > > Best, > Matthew > > _______________________________________________ > AMBER mailing list > AMBER.ambermd.org > > https://nam10.safelinks.protection.outlook.com/?url=https%3A%2F%2Furldefense.com%2Fv3%2F__http%3A%2F%2Flists.ambermd.org%2Fmailman%2Flistinfo%2Famber__%3B!!CzAuKJ42GuquVTTmVmPViYEvSg!M_sE739oXyJHj761ZXLacoC02jUOzCaTzaQ54kOXQHaOH8RCh9a8HiGWGvKWQm6Wj8lxi_CJxGU%24&data=05%7C01%7Cmatthew.guberman-pfeffer%40uconn.edu%7C3bc44cb73bf94ca3abde08dbe646ca54%7C17f1a87e2a254eaab9df9d439034b080%7C0%7C0%7C638356963936851278%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=tSvHld1%2FMxYDxK71pa76vsgwPb1%2F2ld3agr9vGF4lSI%3D&reserved=0 > > _______________________________________________ > AMBER mailing list > AMBER.ambermd.org > > https://nam10.safelinks.protection.outlook.com/?url=http%3A%2F%2Flists.ambermd.org%2Fmailman%2Flistinfo%2Famber&data=05%7C01%7Cmatthew.guberman-pfeffer%40uconn.edu%7C3bc44cb73bf94ca3abde08dbe646ca54%7C17f1a87e2a254eaab9df9d439034b080%7C0%7C0%7C638356963936851278%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=fgM1AUmcqX7uoCfviiGLuE5JI%2B4QbUOK5kkFOBcU4pQ%3D&reserved=0 > > >
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber

(image/png attachment: Screenshot_2023-11-15_at_8.59.22_PM.png)
(image/png attachment: Screenshot_2023-11-15_at_8.59.56_PM.png)
