Date: Thu, 2 Nov 2023 00:31:27 -0700
Hello,
I am interested in computing Histidine pKas. I decided to explore the
Continuous constant pH MD in implicit solvent and implemented a calculation
using this guide by Jack Henderson, Ruibin Liu, Julie Harris, Yandong
Huang, Vinicius Martins de Oliveira, and Jana Shen here:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9910290/, specifically the
GBNeck2-CPHMD simulation in Amber.
However, I am unable to observe a titration curve when titrating at
different pHs. I am doing a range from pH 3.0 to pH 10.0, and doing 10 ns
of simulation at each pH. I am observing that a large portion of lambda
values (nearly 65%) are in "mixed" state, aka 0.2 <= lambda <= 0.8, which
is in contrast to Figure 2 of the paper I linked above. At each pH, the
fraction of protonated/unprotonated is basically 50% so there is no
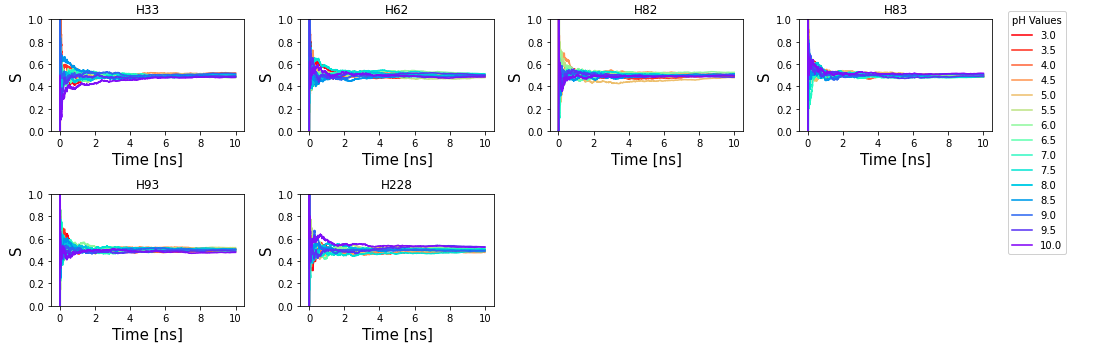
titration curve. I have attached a figure that shows the unprotonated
fraction for 6 his residues in model system 1PTD and all the pHs converge
to 0.5 after a few ns of simulation time. Do I have an error in my
implementation? Why are so many of my lambdas in the mixed state? I have
attached a sample input production script. It seems implausible to me that
my issue can be solved by doing replica-exchange because the values are
converging, they are just converging to 0.5 erroneously at every pH.
*Example production input script:*
Production at pH 4.0
&cntrl
imin=0, nstlim=5000000, dt = 0.002,
irest=1, ntx=5, ig=-1,
tempi=300.0,
temp0=300.0,
ntc=2,
ntf=2,
ntwx=0,
ntwe=0,
ntwr=10000,
ntpr=0,
cut=999,
iwrap=0, igb = 8,
ntt=3, gamma_ln=1.0, ntb=0,
iphmd=1, solvph=4.0, saltcon=0.15,
ioutfm=1, ntxo=2
/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: pH_convergence.png)
