Date: Mon, 3 Jul 2023 17:21:21 +0000
Hi,
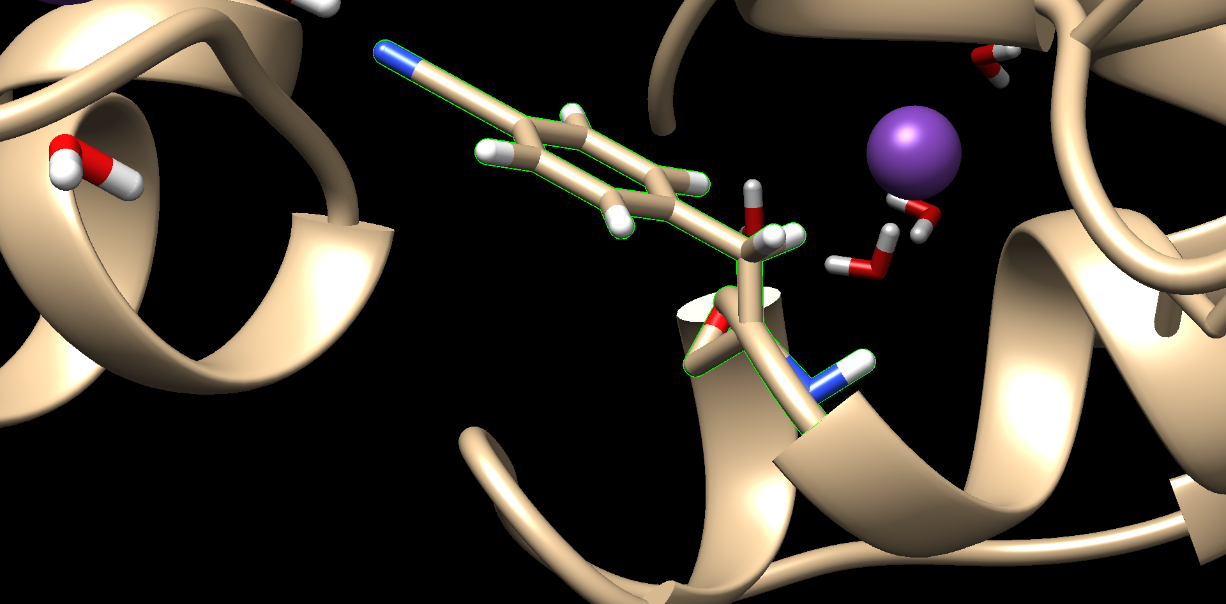
I am currently performing a site-specific mutation on a protein's helix using an unnatural amino acid. During the process of adding ions and water molecules to the system utilizing the tleap program, I observed that the mutated probe is not maintaining its helical structure and is experiencing a disruption or unwinding in the middle. It is important to note that there are no apparent errors in the tleap file. I also have a lib file for unnatural amino acid. I am seeking assistance to understand the underlying cause of this issue and to explore potential solutions to rectify it. Any guidance or insights would be greatly appreciated.
[cid:b873814a-8f95-4b91-8b37-02028cc9bd7b]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Outlook-qvyrojqm.png)
