Date: Wed, 21 Apr 2021 13:37:59 -0700
Hi Kenneth,
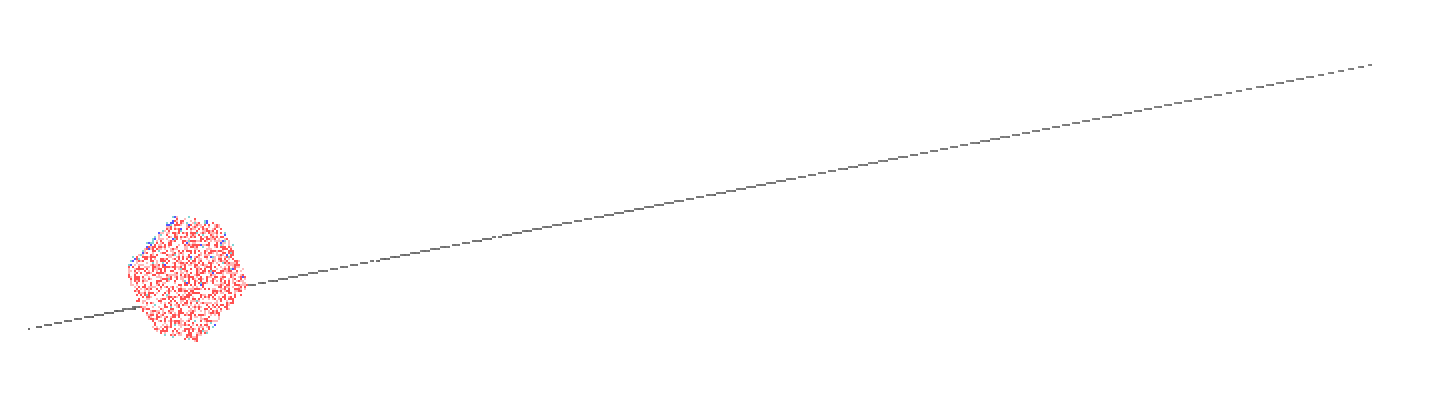
Yes, I have tried that. I've also turned iwrap=0 and the exact same issue
is still there. The area sticking out of the box also distorts locally
before shooting out over 100 nanometers, despite being restrained. The
final distortion is also much larger than the unit cell length (see
attached), so it's not just popping into an adjacent cell.
The energies in the mdout file also spikes right before the protein heavily
distorts.
I have dealt with imaging issues in the past, and I've never seen anything
quite like this.
NSTEP = 2 TIME(PS) = 0.004 TEMP(K) = 6.93 PRESS =
0.0
Etot = -1233660.5161 EKtot = 4504.6196 EPtot =
-1238165.1357
BOND = 876.6350 ANGLE = 3953.7721 DIHED =
6045.6337
UB = 0.0000 IMP = 0.0000 CMAP =
2363.2836
1-4 NB = 7579.1417 1-4 EEL = 78180.4717 VDWAALS =
109000.5146
EELEC = -1446165.1044 EHBOND = 0.0000 RESTRAINT =
0.5163
EAMBER (non-restraint) = -1238165.6520
------------------------------------------------------------------------------
NSTEP = 3 TIME(PS) = 0.006 TEMP(K) = 25.26 PRESS =
0.0
Etot = -1233480.5499 EKtot = 16418.8125 EPtot =
-1249899.3624
BOND = 853.5182 ANGLE = 3654.1325 DIHED =
6041.8623
UB = 0.0000 IMP = 0.0000 CMAP =
2351.2961
1-4 NB = 7507.8397 1-4 EEL = 78040.9065 VDWAALS =
108808.1071
EELEC = -1457163.4803 EHBOND = 0.0000 RESTRAINT =
6.4556
EAMBER (non-restraint) = -1249905.8180
------------------------------------------------------------------------------
NSTEP = 4 TIME(PS) = 0.008 TEMP(K) =********* PRESS =
0.0
Etot = ************** EKtot = ************** EPtot =
-1091938.3276
BOND = 813.7245 ANGLE = 3606.8652 DIHED =
6049.2609
UB = 0.0000 IMP = 0.0000 CMAP =
2334.4278
1-4 NB = 11546.2703 1-4 EEL = 77840.2321 VDWAALS =
278138.1904
EELEC = -1472294.4335 EHBOND = 0.0000 RESTRAINT =
27.1349
EAMBER (non-restraint) = -1091965.4625
------------------------------------------------------------------------------
Date: Tue, 20 Apr 2021 15:32:01 -0400
From: Kenneth Huang <kennethneltharion.gmail.com>
Subject: Re: [AMBER] protein sticking out of solvation box causing
instability
To: AMBER Mailing List <amber.ambermd.org>
Message-ID:
<CALeh7kDVpL-ozdoYbXeFvb1CY4-Lto6+_FR=P1uRzhxLBShL3Q.mail.gmail.com>
Content-Type: text/plain; charset="UTF-8"
Hi,
It looks like an imaging issue- have you tried looking at it after imaging
it with something like
trajin equil.rst
autoimage
trajout test.pdb
Alternatively, I think VMD can draw the other unit cells with an option
under Graphics/Representations, since your protein tail is likely just
sitting in the box next to the orginal.
Best,
Kenneth
On Tue, Apr 20, 2021 at 1:36 PM Neville Bethel <nevillebethel.gmail.com>
wrote:
> Hi everyone,
>
> I came across an issue I haven't seen before.
> I am trying to solvate and equilibrate a large triangular protein.
> tleap seems to put one of the ends of the protein outside the original
> solvation box (trimer_example_0.png attached).
> This seems fine since there is still plenty of space between this end and
> the next periodic image, but when I try to equilibrate, this edge of the
> protein flies off into space (trimer_example_8.png).
>
> I also attached the output text from the equilibration run. Does anyone
> know what's going on here?
>
> Thanks,
> Neville
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screen_Shot_2021-04-21_at_1.25.19_PM.png)
