Date: Sun, 14 Mar 2021 13:45:39 +0530
Dear Users,
I am trying to simulate protein_mrna_sirna complex. I tried to run energy
minimization (using Sander (serial) Amber20) with the input file:
&cntrl
imin=1, maxcyc=20000,ncyc= 10000, cut=10.0, igb=0, ntb=1, ntp=0, ntpr=100,
/
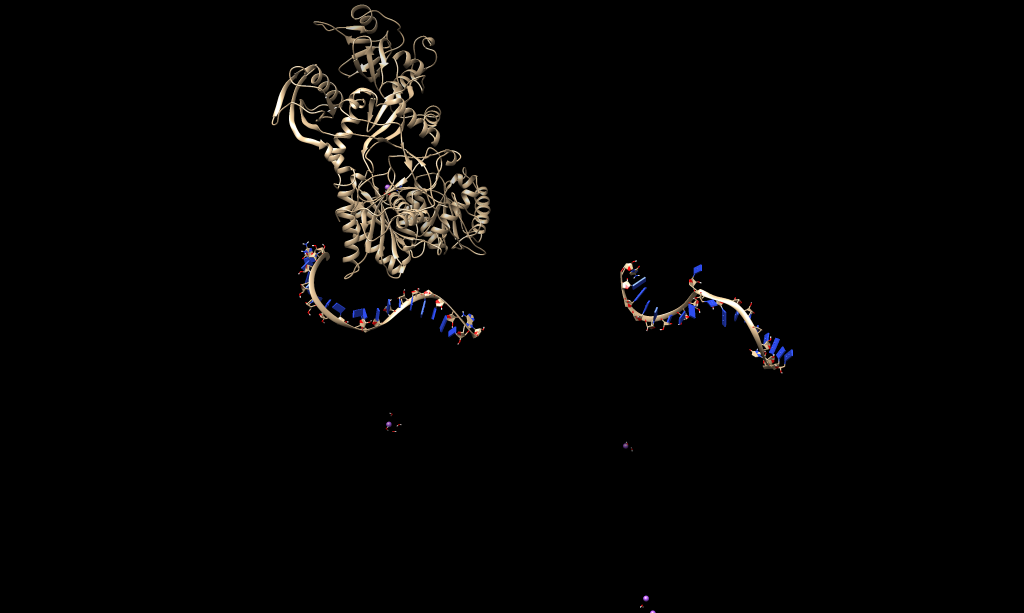
but jast after 200 steps of minimization, protein, sirna and mrna all three
get separated to each other (fig. is attached).
but, if I use ntb=0 then it remain as a complexed form, while ntb=0 meas
simulation in VACUME, while I have to simulate in explicite water.
PLease help me how to retain the complexed structure during energy
minimization. I am not getting where the problem is?
the input tleap is:
source leaprc.protein.ff14SB
source leaprc.RNA.OL3
loadOff terminal_monophosphate.lib
source leaprc.water.tip3p
designed = loadPdb 2032.amber.pdb
check designed
addions designed Na+ 5
solvatebox designed TIP3PBOX 10.00 0.75
charge designed
saveAmberParm designed 2032.trim.prm.top 2032.trim.prm.crd
savepdb designed 2032.out.pdb
quit
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
