Date: Fri, 06 Nov 2020 12:21:48 +0100
Hi Bill,
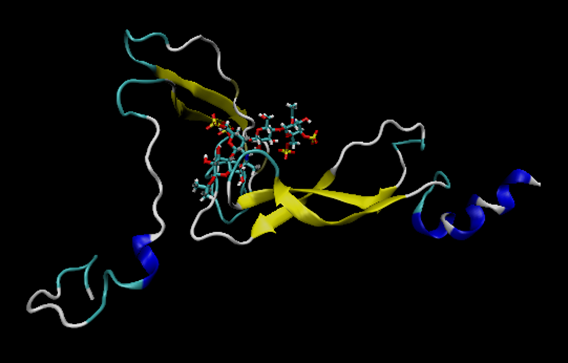
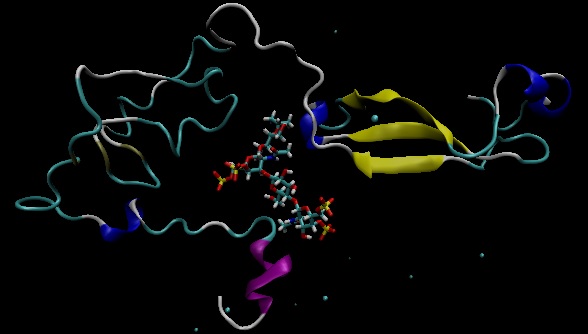
I rectify with my previous message, sorry. I have checked just that,
and indeed my disulfide bond was not defined, and that's why it
appeared broken in all dynamics. So my problem is summarized in: Even
not having put this bond in my input ... because in some cases one of
the two domains of the protein disappears? I am attaching two images
so you can see it better. I have opened the final molecule with
several programs and only one of two domains appears...
Thanks in advance,
Myriam
Bill Ross <ross.cgl.ucsf.edu> escribió:
> Normally bonds that seem to disappear were not made in the first place.
>
> Bill
>
>
> On 11/6/20 2:45 AM, MYRIAN TORRES RICO wrote:
>> Dear all,
>>
>> I wanted to take a query about some results that I have obtained in my
>> molecular dynamics.
>> I have launched a dynamic of a protein-ligand complex (the ligand is a
>> tretasaccharide), and of the two domains that the pdb of my protein
>> initially presents, the disulfide bonds that form one of them ends up
>> breaking ... But the energy data endings are the same as in any other
>> dynamic that has been launched under the same conditions and with the
>> same complex ...
>> Has someone this or something similar happened to him?
>> any ideas?
>>
>> Thanks in advance,
>>
>> Myriam
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
> --
> Phobrain.com
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: smp-196ptn4.jpg)
(image/jpeg attachment: smp-196ptn3.jpg)
