Date: Tue, 29 Sep 2020 01:25:15 -0700
Dear Amber users,
I am trying to calculate PMF for the lipid membrane system and using this
tutorial as a reference (
https://github.com/callumjd/AMBER-Umbrella_COM_restraint_tutorial) with
Amber 18.
I am trying to reproduce some results as described in the tutorial as well
as in this paper (https://doi.org/10.1021/jp903248s), also mentioned in the
tutorial.
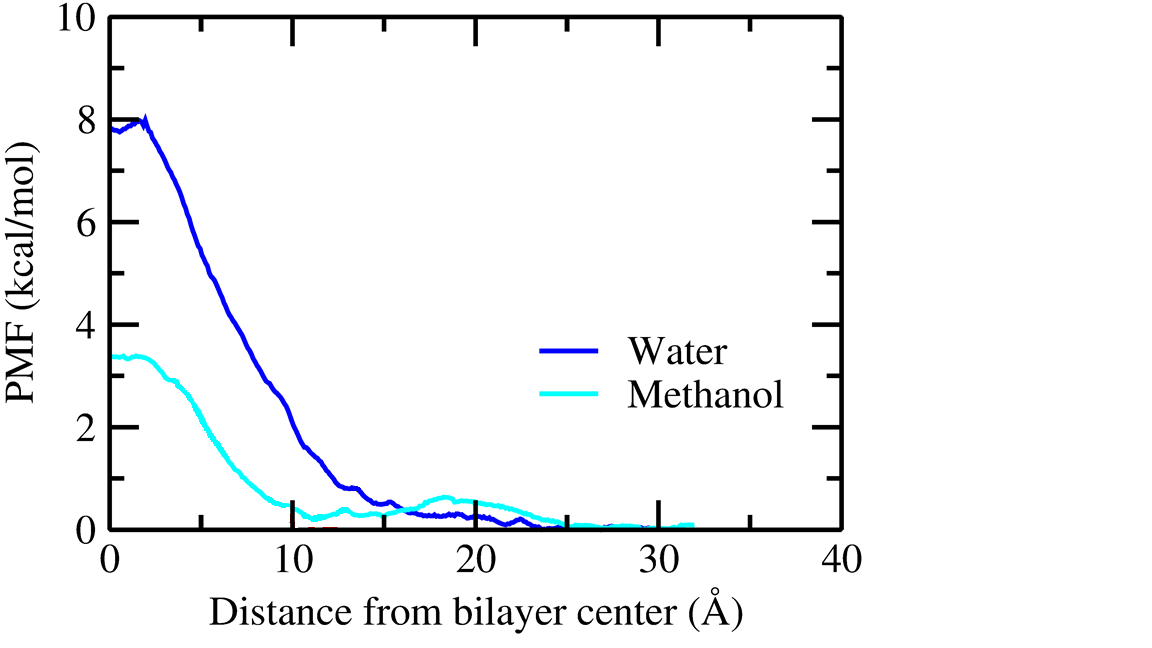
I did it for water and methanol. What I faced that in the case of methanol
there
do not have any single negative value as it should be. I took 0.5 A as
window
width and ran a test simulation of 5 ns for each. I know that it's not
enough for convergence, still, am I missing something? Kindly help me out.
Thanks in advance.
Regards
Somdev Pahari
Research Scholar
NIT Rourkela
Odisha, India
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: PMF.png)
