Date: Mon, 7 Sep 2020 12:42:23 +0300
Hello,
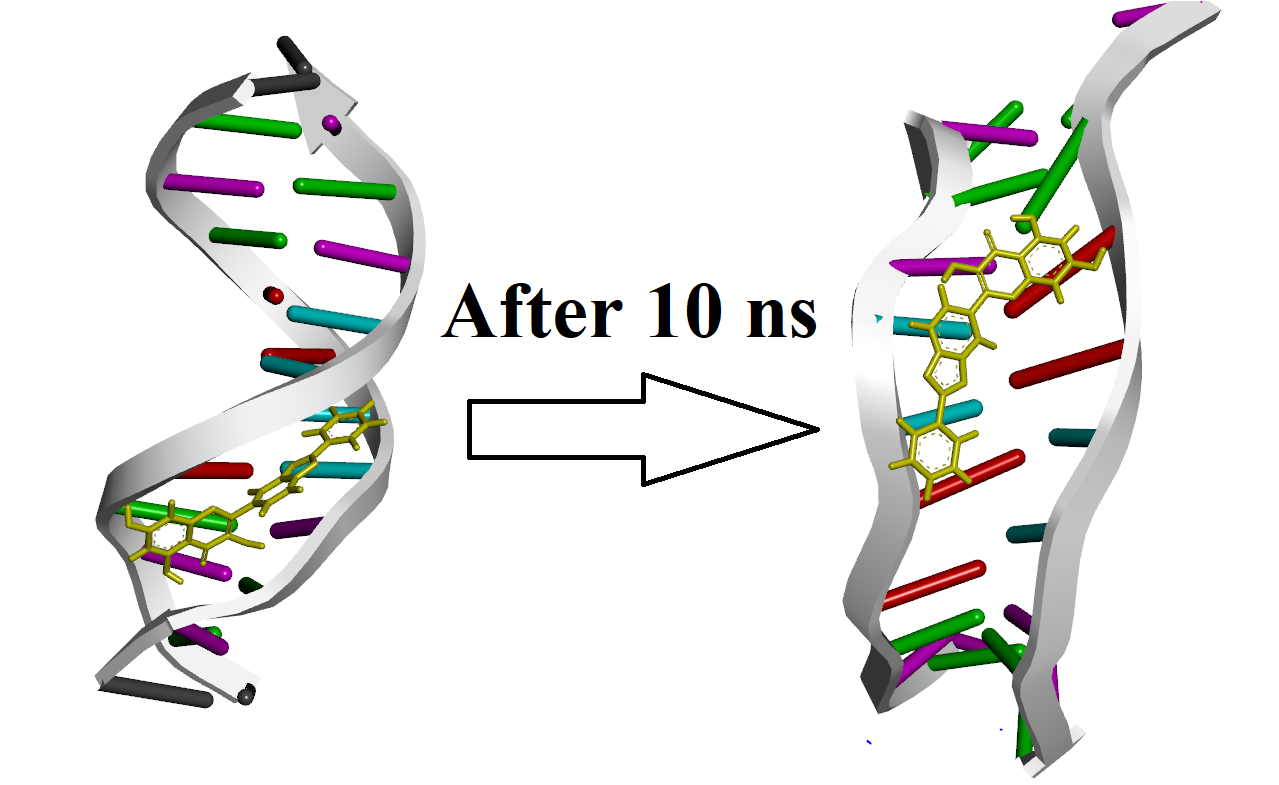
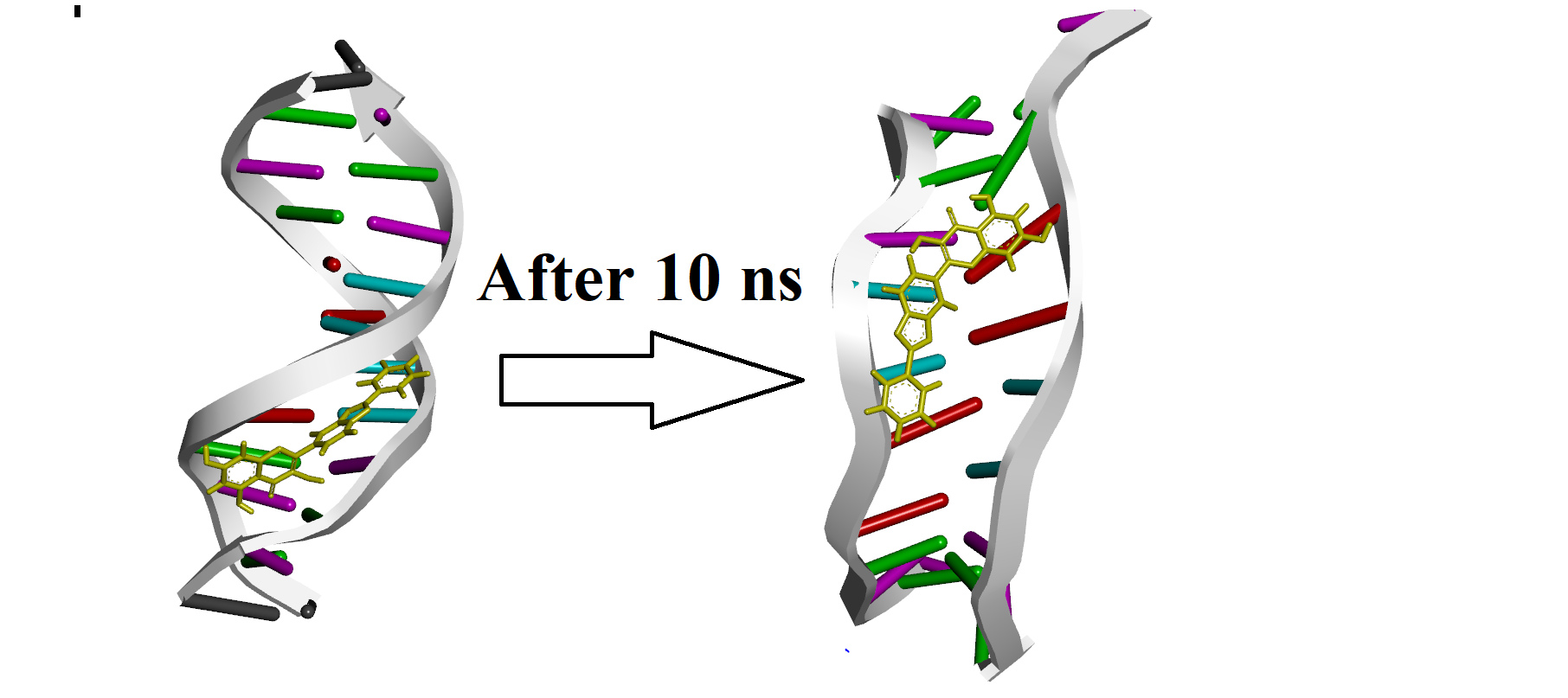
I docked my boron containing a ligand (with boron's force field) with DNA.
After 10 ns it seems ligand is intercalating DNA. Potential energy diagram
seems OK it is very stable. But RMSD increases up to 9A and it is still
increasing (after 10 ns). is it normal or should it be lower? if it should
be lower how can I fix this? I attached first and the last frame of
simulation. if amber members checked and shared their valuable comments I
would be very happy.
here is first and the last frame of simulation:
[image: image.png]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: Untitled.png)
