Date: Tue, 14 Apr 2020 11:24:37 -0400
Does anyone else have this issue when compiling Amber18 with CUDA10.1 and
GCC/8.3.0? Is there any clue how to fix it?
Thanks,
Cong
On Sat, Apr 11, 2020 at 10:25 AM Cong Liu <cong.liu.1.stonybrook.edu> wrote:
> Hi David,
>
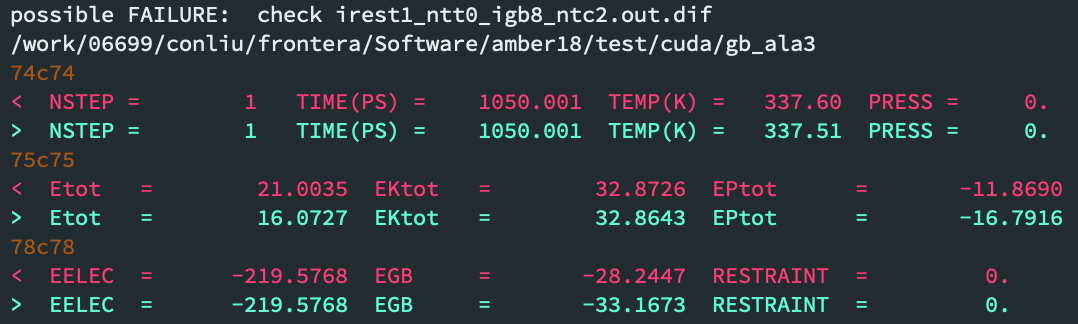
> Thanks for your response. Here is the screenshot of one of the failure:
> [image: Screen Shot 2020-04-11 at 10.22.57.png]
>
> I guess the problem is more serious, since the EPtot and EGB are quite
> different.
>
> Best,
> Cong
>
>
>
> On Sat, Apr 11, 2020 at 8:03 AM David A Case <david.case.rutgers.edu>
> wrote:
>
>> On Fri, Apr 10, 2020, Cong Liu wrote:
>> >
>> >I recently compiled Amebr18 with GCC/8.3.0 and CUDA/10.1. The compilation
>> >went through well. However, *amber_cuda_serial *testing indicates 5
>> "*file
>> >comparison failure*": DPFP myoglobin_igb7, DPFP myoglobin_igb8, DPFP
>> >myoglobin_igb8_gbsa, DPFP irest1_ntt0_igb7_ntc2, DPFP
>> irest1_ntt0_igb8_ntc2
>> >with "IEEE_UNDERFLOW_FLAG IEEE_DENORMAL" floating-point exceptions.
>>
>> The underflow flag is not worrisome, but you should look at the .dif
>> files (collected in the logs folder) and see if the discrepancies look
>> like roundoff errors or something more serious.
>>
>> ....dac
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screen_Shot_2020-04-11_at_10.22.57.png)
