Date: Tue, 20 Aug 2019 22:32:14 -0600
Hello,
Thank you for the quick reply. Please let me know where can I find the
naming templates.
1. By the way, PDB4AMBER generated the TER cards after each NLN, I can
manually delete them.
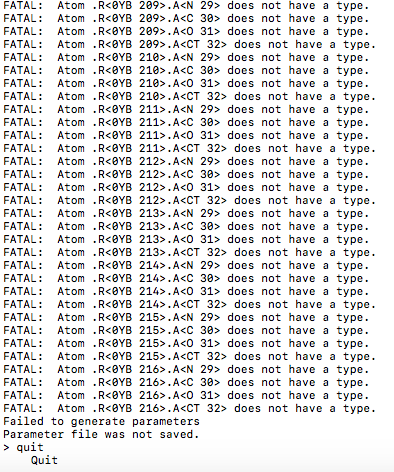
2. NLN are 0YB are the correct naming. But as you can see in my leap input
file, I already source the force field specific for glycans. Now the only
problem is the glycan.
[image: Screen Shot 2019-08-20 at 10.23.18 PM.png]
3. I used PDB2PQR to predict the protonation states of the protein, PDB2PQR
generates the hydrogens. I manually deleted all the hydrogens. Does it
influence the protonation state of the protein?
Thank you so much.
Best regards,
Rui
On Tue, Aug 20, 2019 at 10:07 PM Lachele Foley <lf.list.gmail.com> wrote:
> Hi!
>
> You shouldn't use TER cards after the NLNs unless they are actually at
> the end of an amino acid chain. They are amino acid (modified), and
> don't follow the glycan rules.
>
> Removing hydrogens isn't necessary, but their names need to exactly
> match the templates in the force fields. Folks usually prefer to just
> remove them rather than rename them all. If any of their positions
> are critical, leave those in and make sure the names are right.
> Otherwise, it's easier to delete them. Or rename them all if you
> prefer to do that.
>
> Your NLN and 0YB atom names need to exactly match the templates as
> well. The same hydrogen-naming logic applies, but you must ensure
> that the heavy-atom names match.
>
> Do you know how to find the lib and prep files where the templates are
> (for naming)?
>
> On Tue, Aug 20, 2019 at 11:49 PM Rui Chen <rchen6.ualberta.ca> wrote:
> >
> > By the way, should I remove all the hydrogens before using PDB4AMBER and
> > why? Thank you.
> >
> > On Tue, Aug 20, 2019 at 9:43 PM Rui Chen <rchen6.ualberta.ca> wrote:
> >
> > > Hello,
> > >
> > > I am preparing a PDB file for running LEaP. Since I add glycans using
> > > CHARMM GUI, I rename the glycans to NLN (N-linkages to ASN) according
> to
> > > AMBER manual (LEaP). However, I got error when I run leap. I changed
> the
> > > TER cards, but it still didn't work. I also checked the connections
> between
> > > the carbohydrates and the ASN amino acids using VMD, I couldn't find
> > > anything wrong. The leap input file, log file, pdb file (mentioned in
> leap
> > > input file) are attached.
> > >
> > > Could you please give me some clue about how to solve the problem?
> Looking
> > > forward to hearing from you.
> > >
> > > Sincerely,
> > > Rui
> > >
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
>
>
>
> --
> :-) Lachele
> Lachele Foley
> CCRC/UGA
> Athens, GA USA
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screen_Shot_2019-08-20_at_10.23.18_PM.png)
