Date: Fri, 31 May 2019 20:32:03 +0100
Dear All,
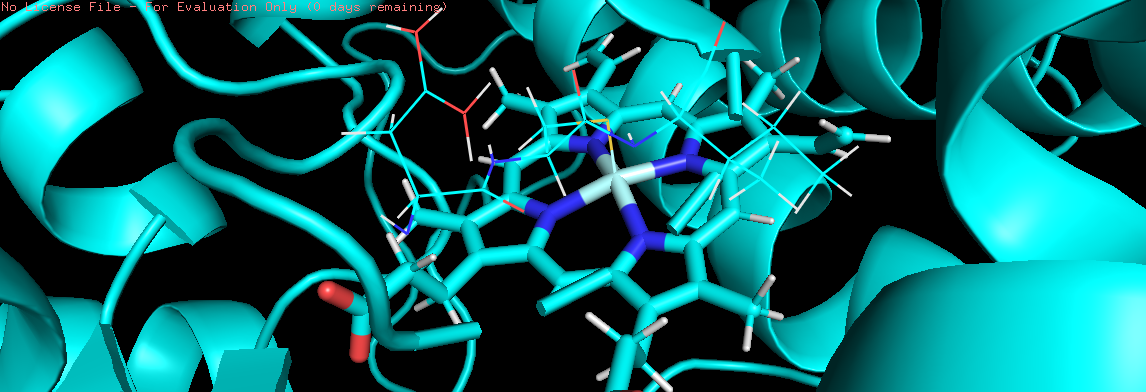
I think the issue is resolved.
I created a pdb with connectivity info from prmtop/inpcrd using ambpdb
command and it shows expected bonds in pymol.
image attached for your reference.
It will be still helpful if someone could explain the meaning of the
message tleap is giving about "*Residues lacking connect*".
I can share the pdb if someone is interested in the structure/topic
(imgage+pdb > 1 MB thus is getting blocked by the admin).
A big thanks to all.
On Fri, May 31, 2019 at 8:05 PM Vaibhav Dixit <vaibhavadixit.gmail.com>
wrote:
> Dear Prof. Case and All,
> thanks for the reminder and your response.
> I move HEM to the end and used pdb4amber to get better PDB file.
> Using this and frcmod shared by Sason Shaik's group, pmrtop/inpcrd are
> generated, but tleap warns about residues not being connected to the main
> chain.
> I think cysteine not being connected to the protein backbone is not
> correct.
> So these prmtop/inpcrd fiels don't represent the protein correctly.
> Did I understand the following tleap message (in *bold*) correctly?
> I would be grateful if HEM parameter experts/authors on the list can
> comment and make suggestions on this issue.
> Thank you all.
> regards
>
> Marking per-residue atom chain types.
>
> * (Residues lacking connect0/connect1 - these don't have chain types
> marked:*
>
> res total affected
>
> * **** * 1
> CASP 1
> CYP 1
> NGLY 1
> NILE 1
> WAT 13994
> )
> (no restraints)
>
> On Fri, May 31, 2019 at 6:30 PM David A Case <david.case.rutgers.edu>
> wrote:
>
>> On Fri, May 31, 2019, Vaibhav Dixit wrote:
>>
>> >Dear Prof. Case,
>>
>> Please send amber-related questions to the mail reflector,
>> amber.ambermd.org,
>> and not to me personally. That way, many people can see your question
>> and try
>> to help, and the answers can help others with similar questions. See
>> http://lists.ambermd.org/mailman/listinfo/amber for information on how
>> to
>> subscribe.
>>
>> In particular, Tom Cheatham and others follow the mailing list, and can
>> probably help.
>>
>> >In another attempt, I used CYX and didn't connect S-Fe.
>>
>> You may have to use the "bond" command in tleap to add missing bonds.
>>
>> >> 1) I removed the atoms causing the error from the PDB, and created
>> bonds
>> >> connecting residues 417-418-419 (since 418 is CYP non-standard
>> cysteine),
>> >> prmtop, inpcrd fails to generate.
>>
>> >> Error: Could not find bond parameter for: C - N3
>> >> Checking for angle parameters.
>> >> Error: Could not find angle parameter: O - C - N3
>> >> Error: Could not find angle parameter: C - N3 - H
>> >> Error: Could not find angle parameter: C - N3 - H
>> >> Error: Could not find angle parameter: C - N3 - H
>> >> Error: Could not find angle parameter: C - N3 - CX
>> >> Error: Could not find angle parameter: CT - C - N3
>>
>> I don't know why these are missing....is there an N3 atom type in CYP?
>> Are you sure there is no frcmod file you are failing to load?
>>
>> In options 2, I wouldn't worry about the ****, but be sure to use parmed
>> to check that you indeed have a bond between the CYP and Fe (as well as
>> other bonds that you need.
>>
>> ...dac
>>
>>
>
> --
>
> Regards,
>
> Dr. Vaibhav A. Dixit,
>
> Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The
> University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
> AND
> Assistant Professor,
> Department of Pharmacy,
> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
> India.
> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>
> ORCID ID: https://orcid.org/0000-0003-4015-2941
>
> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>
> P Please consider the environment before printing this e-mail
>
>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: 1W0E-checkbonds.png)
