Date: Tue, 4 Sep 2018 09:25:25 +0800
Thank you for response.
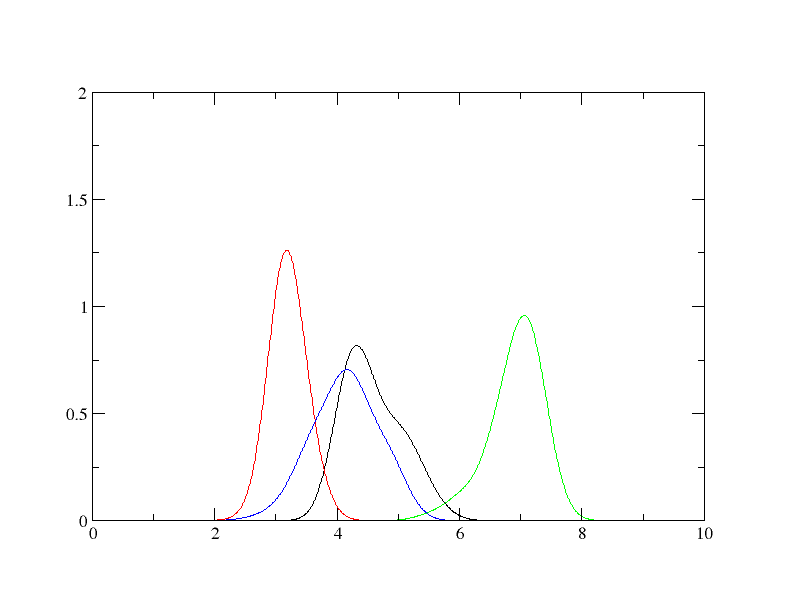
Here's how the figure look like. I used a bandwidth of 0.25.
But if I do not specify the bandwidth, the calculated bandwidth from normal
distribution approximation ranges from 0.02 - 0.05.
I choose 0.25 because I have other systems also to analyzed with the same
dataset and I want to normalize the KDE calculations for all the system I
will be analyzing.
Thank you!
Best,
Kevin
On Mon, Sep 3, 2018 at 9:37 PM Adrian Roitberg <roitberg.ufl.edu> wrote:
> Hi,
>
> Not fully sure how cpptraj computs KDEs, but my guess is that you are
> plotting probability densities, NOT probabilities. Easy to check: If the
> integral of your KDE = 1, then you are plotting probability densities,
> which could be of course larger than 1, depending on the step you chose.
>
> Adrian
>
>
> On 9/1/18 8:48 AM, Daniel Roe wrote:
> > Not sure without actually seeing your data, but I suspect you’re
> choosing a
> > bandwidth that’s too big, leading to underfit data. What is the bandwidth
> > that cpptraj chooses for each (via the normal distribution
> approximation)?
> >
> > -Dan
> >
> > On Fri, Aug 31, 2018 at 11:24 PM Mac Kevin Braza <mebraza.up.edu.ph>
> wrote:
> >
> >> Hello,
> >>
> >> I recently had RMSD values of my protein and use the data from cpptraj
> to
> >> do KDE analysis )also in cpptraj). However, I am getting values greater
> >> than one at specific bandwidth. Is there any way I can avoid getting
> >> probability values > 1 at fixed bandwidth.
> >>
> >> This is how my cpptraj input file look like:
> >>
> >> readdata sotyr_apo_EL1_rmsd.dat name EL1
> >> readdata sotyr_apo_EL3_rmsd.dat name EL3
> >> readdata sotyr_apo_IL3_rmsd.dat name IL3
> >> readdata sotyr_apo_IL4_rmsd.dat name IL4
> >>
> >> kde EL1 out SoTyrR_EL1_kde.out min 0 max 10 step 0.01 bandwidth 1
> >> kde EL3 out SoTyrR_EL3_kde.out min 0 max 10 step 0.01 bandwidth 1
> >> kde IL3 out SoTyrR_IL3_kde.out min 0 max 10 step 0.01 bandwidth 1
> >> kde IL4 out SoTyrR_IL4_kde.out min 0 max 10 step 0.01 bandwidth 1
> >>
> >> run
> >> quit
> >>
> >> Thank you very much!
> >>
> >> Best regards,
> >> Mac Kevin E. Braza
> >> _______________________________________________
> >> AMBER mailing list
> >> AMBER.ambermd.org
> >>
> https://urldefense.proofpoint.com/v2/url?u=http-3A__lists.ambermd.org_mailman_listinfo_amber&d=DwIGaQ&c=pZJPUDQ3SB9JplYbifm4nt2lEVG5pWx2KikqINpWlZM&r=JAg-KQEjdZeg_E8PHDDoaw&m=wliBR8JmmkJM7QlgORe2pDmfh-Qz2iY0KNoHSw0U-WA&s=yqm4Bsfhsp5tRZUTqV_z5qLzWQITNzhJGW5xzxgukow&e=
> >>
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> >
> https://urldefense.proofpoint.com/v2/url?u=http-3A__lists.ambermd.org_mailman_listinfo_amber&d=DwIGaQ&c=pZJPUDQ3SB9JplYbifm4nt2lEVG5pWx2KikqINpWlZM&r=JAg-KQEjdZeg_E8PHDDoaw&m=wliBR8JmmkJM7QlgORe2pDmfh-Qz2iY0KNoHSw0U-WA&s=yqm4Bsfhsp5tRZUTqV_z5qLzWQITNzhJGW5xzxgukow&e=
>
> --
> Dr. Adrian E. Roitberg
> University of Florida Research Foundation Professor
> Department of Chemistry
> University of Florida
> roitberg.ufl.edu
> 352-392-6972
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: KDE_sample.png)
