Date: Fri, 31 Aug 2018 20:04:10 +0000
Hi,
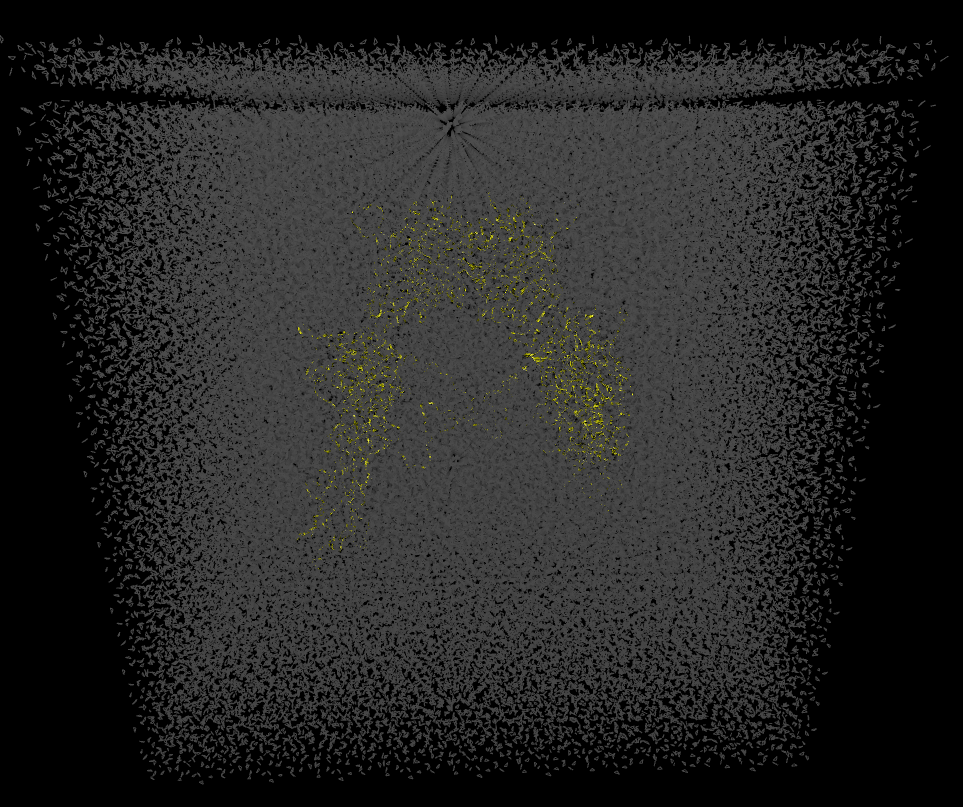
Thank you for the help. For the cellulose system, it workd well after I added the “anchor” command into the input file. But for my protein system, cpptraj will still move the bottom layer to the top, and there will be a gap in the water box. The topology(.parm) and restart(.rst) files was generated from the xleap. Attached figure shows the system after the cpptraj. What do you think the problem might be? Do you think it is problem with my cpptraj command or xleap command?
Tianyi
________________________________
From: Daniel Roe <daniel.r.roe.gmail.com>
Sent: Thursday, August 30, 2018 8:59 PM
To: AMBER Mailing List
Subject: Re: [AMBER] How to use cpptraj to wrap protein
Hi,
By default, autoimage assumes the first molecule of the system should
be the "anchor" molecule, i.e. the molecule in the center of the
system. In this case that is not true; the first molecule is on the
outside of the cellulose bundle. You need to pick one near the center
of the bundle. When I look at the cellulose bundle it appears that
molecule 50 (residues 2010 to 2050) is close to the center, so
something like:
# Using new molecule syntax
parm prmtop
trajin inpcrd
autoimage anchor ^50
trajout out.crd
will work. If you're using an old version of cpptraj that does not
support molecule syntax you can substitute ^50 with :2010-2050, but I
recommend upgrading to the latest version.
Hope this helps,
-Dan
On Wed, Aug 29, 2018 at 8:12 PM Yang, Tianyi <TiaYang.clarku.edu> wrote:
>
> Hello:
>
> I am learning how to use cpptraj of Amber 16 to wrap the protein and water into the box. I am using the cellulose system from the Amber benchmark. The topology, restart and input files were downloaded from the Amber benchmark website. After cpptraj, water molecules were wrapped back into the box but the protein was not in the center. And it seems that the bottom of the box was moved to the top.
>
>
> My cpptraj input(rst.cpptraj file):
>
> trajin inpcrd
>
> autoimage
>
>
> cpptraj command:
>
> $AMBERHOME/bin/cpptraj -p prmtop -i rst.cpptraj &>rstcpp.log -x out.crd
>
>
> Is there anything wrong with my cpptraj command or input file? The picture shows the system after the cpptraj.
>
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
AMBER -- AMBER Mailing List<http://lists.ambermd.org/mailman/listinfo/amber>
lists.ambermd.org
AMBER -- AMBER Mailing List About AMBER: This is the AMBER Mailing List. It is designed to provide a forum for users of the AMBER Molecular Dynamics and related software to ask questions related to AMBER and Molecular Dynamics Simulations in general.
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: cpptarj.png)
