Date: Fri, 10 Aug 2018 14:08:41 +0200
Dear Amber Users,
I have run PCA analysis of my system, in which I think there are few
different conformational states.
This is the cpptraj I have used:
parm prot_A_c0_HH_2_FS.prmtop
trajin trajectory_1_c0.nc
trajin trajectory_2_c0.nc
trajin trajectory_3_c0.nc
trajin trajectory_4_c0.nc
trajin trajectory_5_c0.nc
rms All :1-394 out rmsd_all.agr mass
average crdset all-sim-c0
createcrd all-sim-traj-c0
run
crdaction all-sim-traj-c0 rms ref all-sim-c0 :1-394
crdaction all-sim-traj-c0 matrix covar \
name all-sim-covar-c0 :1-394
runanalysis diagmatrix all-sim-covar-c0 out all-sim-c0-evecs.dat \
vecs 3 name myEvecs \
nmwiz nmwizvecs 3 nmwizfile prot.c0.nmd nmwizmask :1-394
crdaction all-sim-traj-c0 projection All modes myEvecs \
beg 1 end 3 :1-394 out Project.modes.c0.dat
hist All:1 All:2 bins 100 out all-sim-c0_hist_1_2.gnu norm name All_1_2
hist All:2 All:3 bins 100 out all-sim-c0_hist_2_3.gnu norm name All_2_3
hist All:1 All:3 bins 100 out all-sim-c0_hist_1_3.gnu norm name All_1_3
run
clear all
parm prot_A_c0_HH_2_FS.prmtop
trajin trajectory_1_c0.nc
trajin trajectory_2_c0.nc
trajin trajectory_3_c0.nc
trajin trajectory_4_c0.nc
trajin trajectory_5_c0.nc
readdata all-sim-c0-evecs.dat name Evecs
runanalysis modes name Evecs trajout all-sim-mode1.c0.nc \
pcmin -100 pcmax 100 tmode 1 trajoutmask :1-394 trajoutfmt netcdf
runanalysis modes name Evecs trajout all-sim-mode2.c0.nc \
pcmin -100 pcmax 100 tmode 2 trajoutmask :1-394 trajoutfmt netcdf
runanalysis modes name Evecs trajout all-sim-mode3.c0.nc \
pcmin -100 pcmax 100 tmode 3 trajoutmask :1-394 trajoutfmt netcdf
projection MyProject evecs Evecs out Project.c0.dat beg 1 end 3 :1-394
filter MyProject:2 min -25 max 5 MyProject:3 min -25 max -10 out
filtered.pc1.pc2
trajout filtered.pc1.pc2.nc NetCDF
run
quit
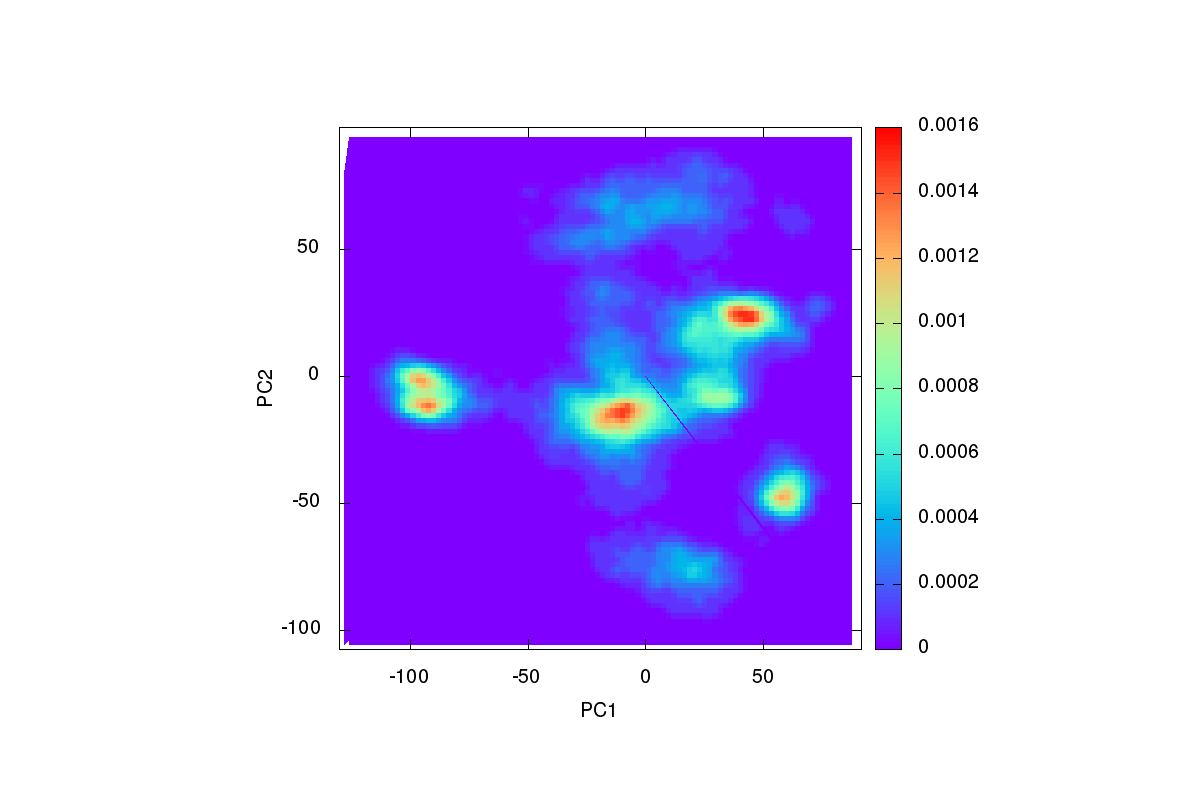
So now I have the distribution of components plots (PC1vsPC2 attached) and
the filtered.pc1.pc2 file, but I still don't know how to find the
structures of interests.
The filtered file looks like this:
#Frame Filter_00004
1 0
2 1
3 0
4 0
5 0
6 1
7 0
8 0
9 0
10 1
...
so I will probably find the structures that fit to the min and max values,
but where can I find the density info?
Is it in the all-sim-c0_hist_1_2.gnu?
Thank you for your patience and help :)
Best regards
Karolina Mitusińska
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: PC1vsPC2_c0.png)
