Date: Sun, 15 Jul 2018 22:04:33 +0000 (UTC)
Thanks a lot for your thoughts, Dan!
It may very well be that longer simulation time would improve the values. What still does not fit is the fact that the MSD plots are not monotonously increasing. The displacement in whatever direction happens should increase the MSD value, don't you agree? I am particularly puzzled by the spike downwards at 50ns - as if the molecule would appear suddenly at the different place - I imagine a wrapping artefact would look like that....
Nevertheless, I checked the water diffusion too: for all water the MSDs are very linear (attached), but for a single water the same non-monotonous behaviour is observed (second figure attached). I am wrong expecting a monotonously increasing MSD plot even for a single molecule?
Any input is appreciated!Kris
On Saturday, July 14, 2018, 6:42:40 PM GMT+2, Daniel Roe <daniel.r.roe.gmail.com> wrote:
Hi,
I think the issue here is that you just do not have enough sampling.
The Einstein relation is:
2*N*D = lim{t->inf}( <MSD> / t)
The key is that this holds true as 't' approaches infinity. The reason
relatively short simulations are sufficient for calculating the
translational diffusion of e.g. water is that you typically have a lot
of them (maybe tens of thousands), and they tend to travel a lot
compared to other molecules in a system (like e.g. solute or lipids).
Over the course of a simulation, all the water may travel on average
an order of magnitude farther than other molecules. In contrast,
because you typically have fewer solute atoms (in your case I'm
guessing maybe 300ish?) and they aren't moving around as quickly you
have much less idea about the overall diffusive behavior over the same
time scale. To convince yourself of this, you can calculate the
diffusion of all the water in your system and compare that to
calculating the diffusion of just a handful of water molecules (or
even just 1). The MSD vs time for all the water should look nice and
nearly straight, while the MSD vs time for the smaller number of
waters should look more rough (and more like the plot you show).
So to summarize, you need to run longer. Also, if you are in fact
calculating diffusion of lipids in a bilayer (seems like too few
molecules here, but just in case) there are some extra considerations
(like box size effects) - see e.g.
https://pubs.acs.org/doi/full/10.1021/acs.jpcb.6b09111 and related
work. Hope this helps,
-Dan
On Thu, Jul 12, 2018 at 6:44 PM, Kris Feher <kris.feher.yahoo.com> wrote:
> Dear All,
> I have bring up a topic that is widely discussed on the list in the past, yet I do not seem to get it right. I would like derive translational diffusion coefficient from Mean Squared Displacements. The simulations were run with iwrap =1 (see protocol below), I processed the trajectories in cpptraj with unwrap and tried to get the diffusion coefficient (second script). I was expecting that the plots the MSDs (overall, total, x,y,z) should be monotonously increasing, but this is not the case (see attached). At around 50ns there is a clear downward spike in all plots, which does not make sense. Accordingly the fitted diffusion coefficients are sort of random (certainly do not match the experimental data) and in some cases are even negative. Not using the noimage keyword does produce slightly different MSD plots with the same issues. Could anyone help me figuring out where it goes wrong?
> Thanks for any idea!Kris
>
>
>
>
> Production
> &cntrl
> imin=0, ntx=5, irest=1,
> ntb=2,cut=8.0,
> ntp=1, pres0=1.0,
> ntc=2,
> ntf=2,
> ntt=3, temp0=310.0, gamma_ln=1.0,
> nstlim=5000000, dt=0.002,
> ioutfm=1,
> iwrap=1, ioutfm=1,
> ntpr=5000, ntwr=50000, ntwx=5000,
> ntr=0, ig=-1,
> /
>
> - cpptraj script for trajectory processing
> parm ../../dna_w.prmtop
> trajin ../../5_prodNPT/dna_prod1.netcdf
> trajin ../../5_prodNPT/dna_prod2.netcdf
> trajin ../../5_prodNPT/dna_prod3.netcdf
> trajin ../../5_prodNPT/dna_prod4.netcdf
> trajin ../../5_prodNPT/dna_prod5.netcdf
> trajin ../../5_prodNPT/dna_prod6.netcdf
> trajin ../../5_prodNPT/dna_prod7.netcdf
> trajin ../../5_prodNPT/dna_prod8.netcdf
> trajin ../../5_prodNPT/dna_prod9.netcdf
> trajin ../../5_prodNPT/dna_prod10.netcdf
> unwrap
> diffusion :1-20 separateout diffusion.dat noimage diffout DiffusionConstants.lst
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
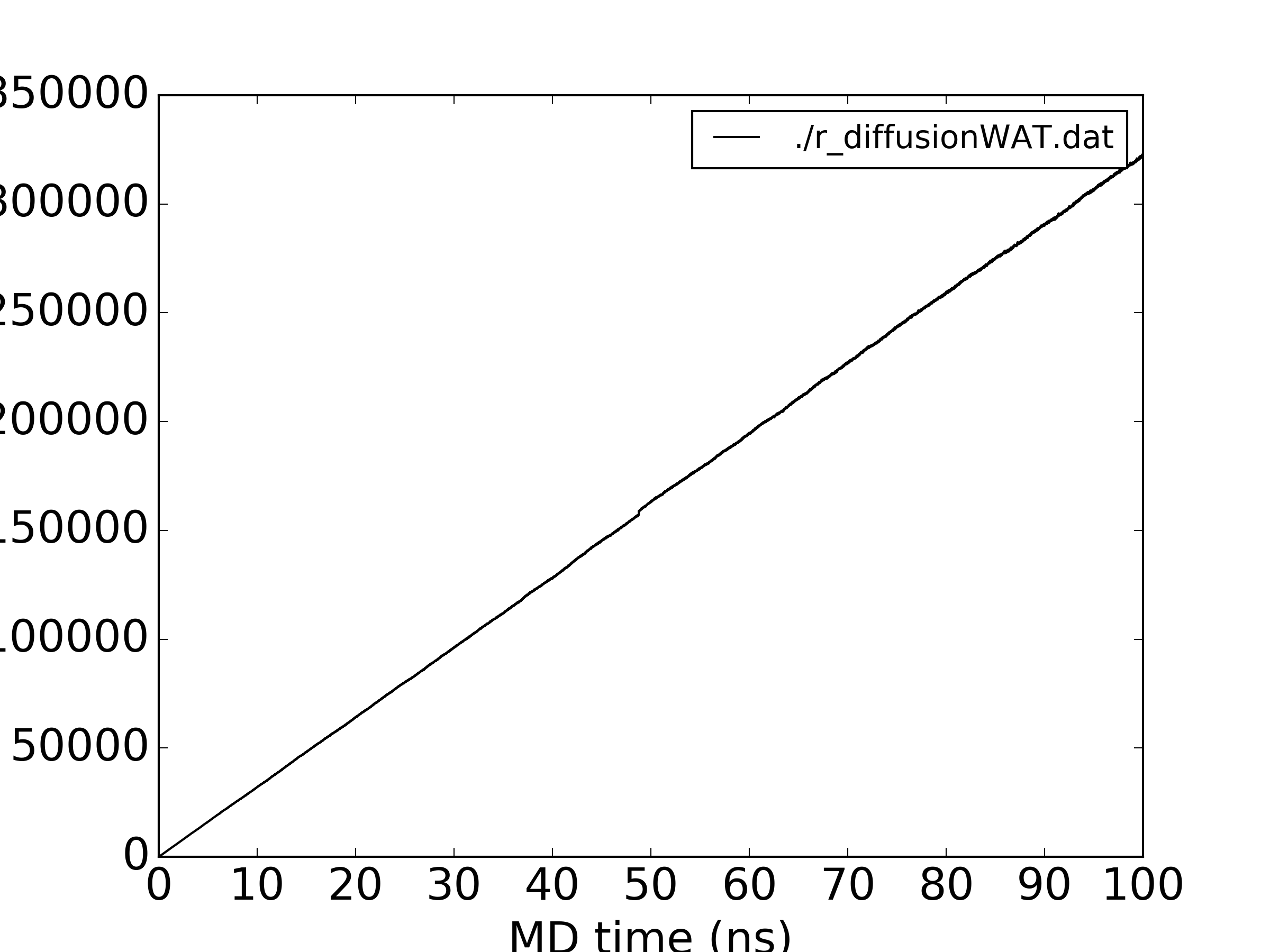
(image/png attachment: r_diffusionWAT.png)
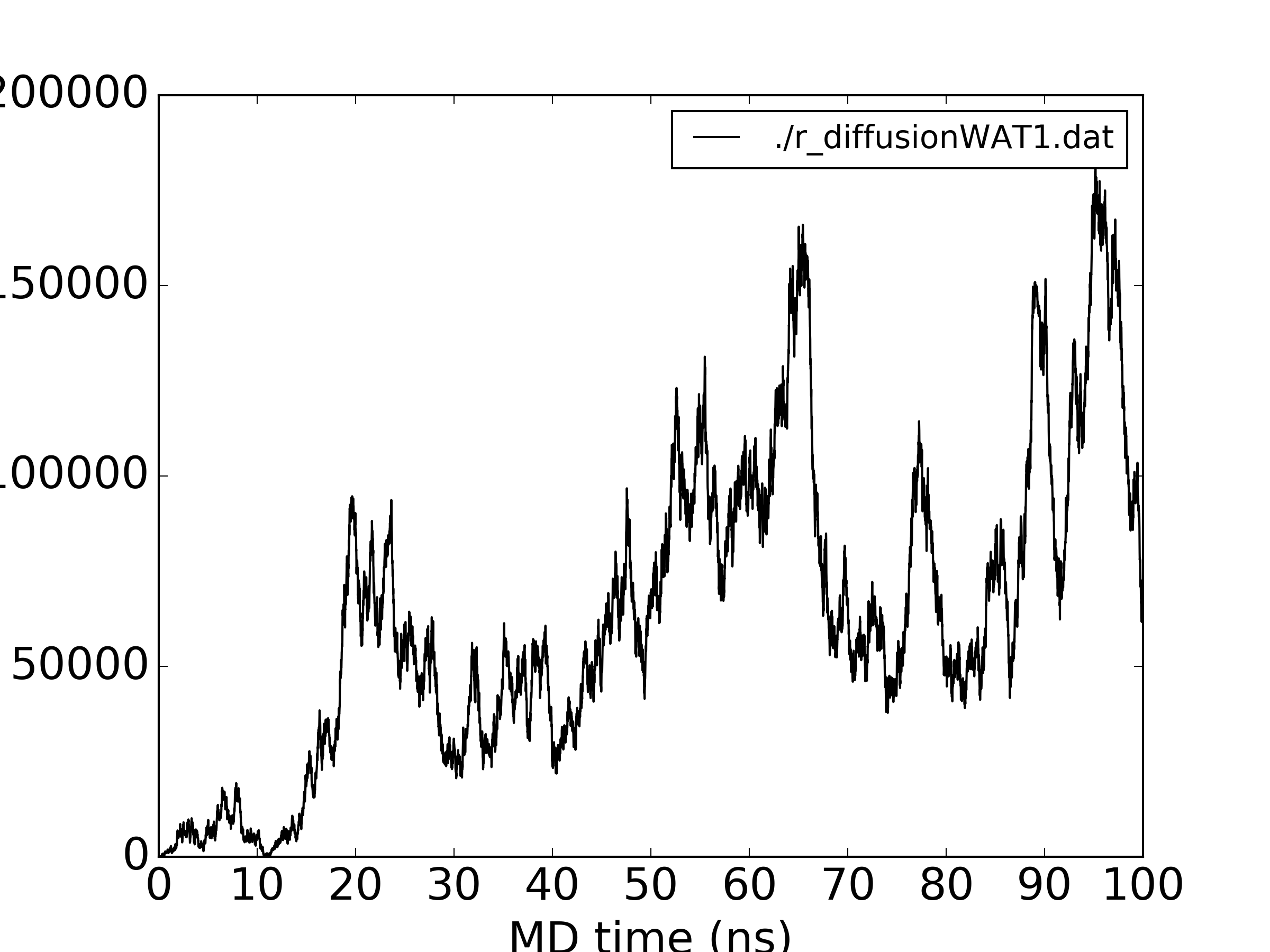
(image/png attachment: r_diffusionWAT1.png)
