Date: Mon, 9 Apr 2018 21:54:27 +0000
Hi everyone,
I am making oxidized and reduced form of amino acids by making them heteroatom. But after making a parameter file when I am visualizing the structure it looks like oxidized/reduced amino acid is not a part of protein. It is a separate ligand (attached image before.png and after.png)
If I remove the TER before heteroatom it is not giving any problem. But amber automatically add ter after heteroatom.
Please suggest me what should I do.
Thanking you,
-Abhishek
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
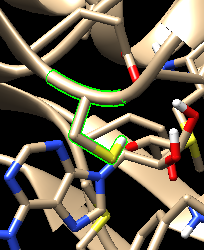
(image/png attachment: before.png)
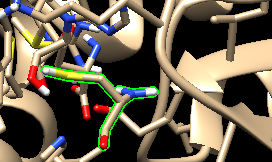
(image/png attachment: after.png)
