Date: Thu, 14 Sep 2017 14:51:00 +0200
Dear Amber users,
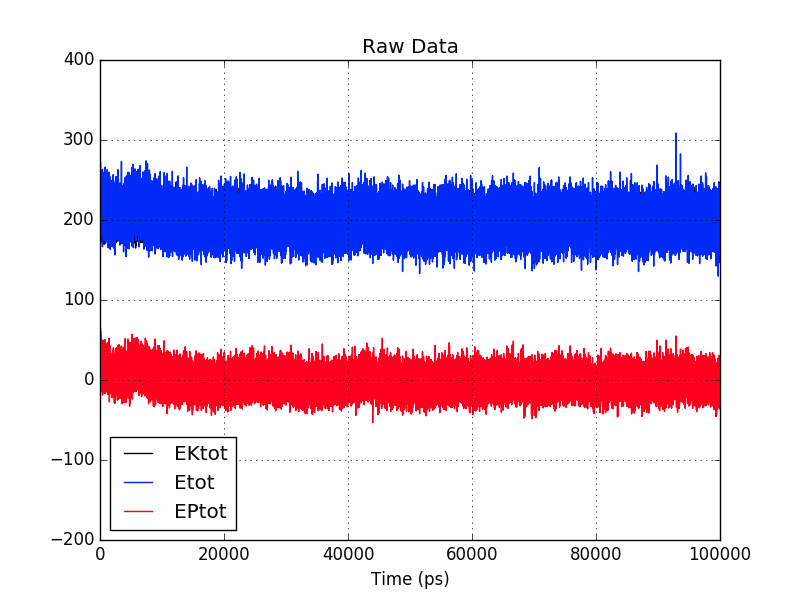
I simulated a peptide for 100 ns and upon mdout_analyzer.py analysis found
that the Etot (Total energy) remains positive and equal to EKtot (kinetic
energy). This was an implicit simulation. The resultant structure is
expected and no problems arose there.
Of course that means that the potential energy of the system (EPtot) did
not contribute to the energy of the system and it almost remains at zero
throughout.
The kinetic energy and total energy overlap each other in the graph
attached below.
The energy remains positive all throughout. Is it normal?
One more detail, the structure was modeled from the scratch using AMBER so
every secondary structure formation happened during the course of 100 ns.
Please advice.
Thank you.
[image: Inline image 1]
[image: Inline image 2]
-- Best wishes Chetna
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber

(image/png attachment: image.png)
(image/png attachment: 02-image.png)
