Date: Tue, 27 Oct 2015 12:05:38 +0000
Hi Adrian,
Many thanks for your comments; they were very useful. Regarding ig==-1, I
am using it except in the single input file I copied in the email. I am now
running the simulation using the generalized distance coordinate as the
difference between the lenghths of the bond to be formed and the one to be
broken with 0.2 A windows. This is my restraint file:
iat=1937,6803,6803,6798, r1=-10.0, r2=1.5, r3=1.5, r4=10.0, rstwt=1,-1,
rk2=50, rk3=50,
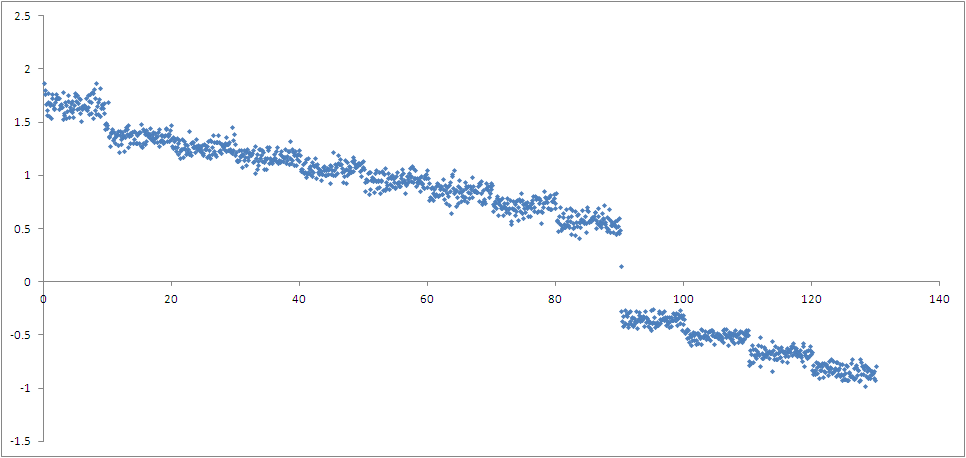
and the overlap has greatly improved. The problem starts to happen when the
coordinate gets closer to the formation of the bond where a big gap appears
in the graph and the coordinate jumps from 0.15 to -0.3 A. I tried
increasing the force near the region but it did not work. This is how the
reaction coordinate versus time looks like:
[image: Inline images 2]
Any advice please
Best Regards,
Mahmood
On 23 October 2015 at 14:39, Adrian Roitberg <roitberg.ufl.edu> wrote:
> Hi
>
> Several comments.
>
> Never run the WHOLE set of simulations before you are satisfied with the
> overlap. Just run 3 or 4 windows and then see if you need to change
> something before going further.
>
> As for your qeustions:
>
> Is that sufficient overlap? If it is not, do I need to reduce the spacing
> or increase the force constant to get better overlap?
>
> You do not show the overlap, so it is hard to tell you, but my guess
> from your first figure is that no, it is not enough. By the way, if you
> want to increase overlap, you need either a closer spacing and/or a
> LOWER force constant(not increased!).
>
> PLEASE add ig=-1 to your mdin !
>
> As for your coordinate, it is NEVER a good idea to use just one distance
> for a reaction. Look at published work on this, and you will see that
> peopel tend to use a reaction coordinate that has RC=distance(bond being
> broken) - distance(bond being made)
>
> That produced much more stable surfaces.
>
> adrian
>
>
>
> On 10/23/15 7:56 AM, Mahmood Jasim wrote:
> > Hi AMBER users,
> >
> > I am trying to simulate a reaction involving the formation of a covalent
> > bond using umbrella sampling combine with QM treatment of the reaction
> > atoms. I have the SG atom of a cysteine residue attacking an electrophilc
> > carbon on a ligand. The distance between these 2 atoms is the reaction
> > coordinate. The windows of umbrella sampling are spaced 0.3 Angstrom
> apart.
> > The restraint file looks like this:
> >
> > #
> > # 123 CYM SG 432 UNK C1 3.43
> > &rst
> > ixpk= 0, nxpk= 0, iat=1937,6803, r1= 2.93, r2= 3.43, r3= 3.43, r4=
> 3.93,
> > rk2=0.0, rk3=100.0, ir6=1, ialtd=0,
> > &end
> >
> > and the md.in file looks like this:
> >
> > &cntrl
> > imin = 0,
> > irest = 1,
> > ntx = 7,
> > ntb = 2,
> > cut = 12,
> > ntr = 0,
> > ntc = 1,
> > ntf = 1,
> > igb = 0
> > ntp = 1
> > tempi = 300.0,
> > temp0 = 300.0,
> > ntt = 3,
> > gamma_ln = 1.0,
> > nstlim =10000, dt = 0.001
> > ntpr = 1000, ntwx = 1000, ntwr = 1000, nmropt = 1,
> > ifqnt=1,
> > /
> >
> >
> > &qmmm
> >
> > qmmask =
> > '.1934-1937,6794-6848',
> > qmcharge =
> > 0,
> > qm_theory =
> > 'PM3',
> > qmcut =
> > 12.0,
> > /
> > &wt type='DUMPFREQ', istep1=100,/
> > &wt type='END' /
> > DISANG=RST-1.dist
> > LISTOUT=POUT
> > DUMPAVE=DIST_1.dat
> >
> > The run was able to simulate the formation of the bond at the end and the
> > next graph represents the values of the coordinates across the windows.
> > [image: Inline images 1]
> >
> > Is that sufficient overlap? If it is not, do I need to reduce the spacing
> > or increase the force constant to get better overlap?
> >
> > I used the WHAM code by Alan Grossfieldto calculate PMF with this
> command:
> >
> > wham 1.59 3.73 70 0.00001 300 0 meta.dat result.dat
> >
> > and this was the meta.dat file:
> >
> > DIST_1.dat 3.43 200
> > DIST_2.dat 3.13 200
> > DIST_3.dat 2.83 200
> > DIST_4.dat 2.53 200
> > DIST_5.dat 2.23 200
> > DIST_5.dat 1.93 200
> > DIST_7.dat 1.63 200
> >
> > and the histogram I am getting looks like:
> >
> > [image: Inline images 2]
> >
> > I was expecting the histogram to look different, the PMF to go higher and
> > then decreases as it approcahes the products. If I managed to improve the
> > overlapping, will this change the histogrm?
> >
> > Many thanks,
> > Mahmood Jasim
> > Aston University
> > UK
> >
> >
> >
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
>
> --
> Dr. Adrian E. Roitberg
> Professor.
> Department of Chemistry
> University of Florida
> roitberg.ufl.edu
> 352-392-6972
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
