Date: Mon, 5 Oct 2015 14:53:49 +0700
Dear Lachele,
Thank you very much for your advice. I really appreciate it.
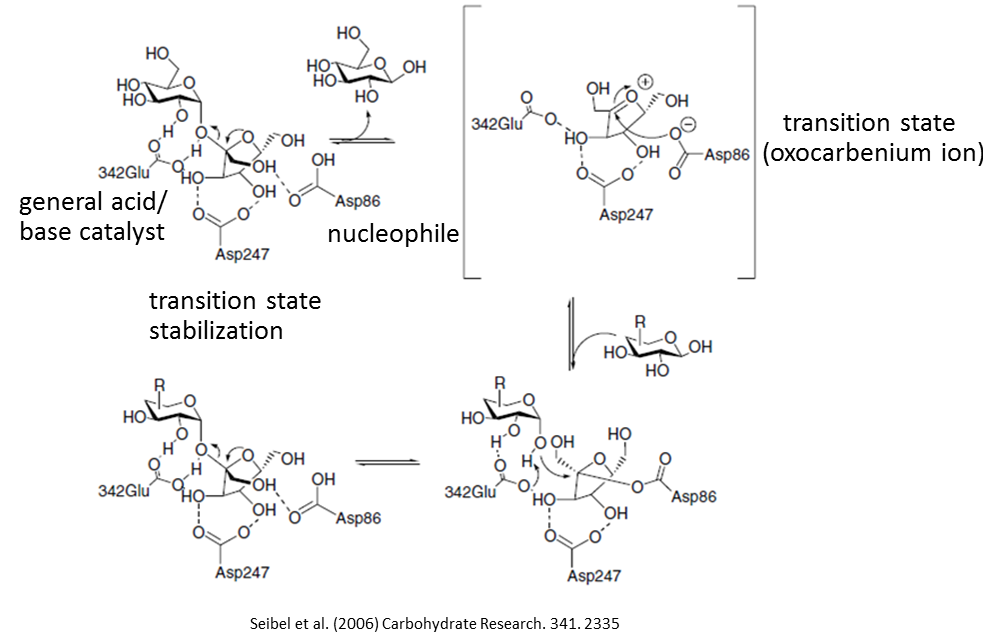
We are trying to build a structure that is a part of the proposed reaction
pathway of levan synthesis (as shown in the attached picture) and to dock
levan of various chain lengths in the active site to gain an understanding
on how levansucrase perform the levan synthesis. I think there are two
possibilities for the models.
1. The structure with Asp86 attached to fructosyl unit.
2. The structure with fructosyl unit as an oxocarbenium ion with no
attachment to Asp86.
For model 1, I’m not sure if it’s possible to use the remove and bond
command in tleap to build a bond between Asp86 and the fructosyl unit
(aspartic acid has ASP and ASH). I’m also not sure how to assign the
charges at the connecting points for this structure.
For model 2, there is no connecting point between Asp86 and the
oxocarbenium ion but I’m not sure if it’s possible to use tleap to build
oxocarbenium ion based on the crystal structure of sucrose in the active
site and how to assign its charges.
I’m wondering if you have any suggestions on how I should proceed? Which
model can be constructed using AMBER (tleap)? What would be the most
appropriate way to assign the charges to the model?
Thank you very much for your help.
Regards,
Surasak
On Sun, Oct 4, 2015 at 11:17 AM, Lachele Foley <lf.list.gmail.com> wrote:
> We do not yet have the capability to link to anything other than ASN,
> THR or SER on the website. We have parameters also for proline, but
> the website doesn't support it. We only make available on the website
> structures for which we have parameters that have been validated for
> that purpose. We are considering allowing builds of other systems,
> but we would not be able to provide files necessary to run a
> simulation. It takes a bit of work to ensure that the parameters are
> made properly.
>
> Ignoring that entirely, I downloaded 1pt2 and took a look at that
> residue in context. It is quite buried in that crystal structure.
> Because of this, you should proceed very carefully when attaching
> anything to it: the forces the structure would be under during
> minimization could easily cause severe distortion of the protein or
> the sugar. If that site is naturally glycosylated, then it seems
> likely that either the protein distorted during the crystallization
> process or the glycosylation occurs before the protein is completely
> folded.
>
>
>
>
> On Fri, Oct 2, 2015 at 12:28 AM, Surasak Chunsrivirot
> <chunsrivirot.gmail.com> wrote:
> > Hi,
> >
> > I want to build a structure of levansucrase (1PT2) with a fructosyl
> residue
> > attached to Asp86 as in the attached picture. I tried the glycoprotein
> > builder website but it doesn't allow me to choose Asp86 as my point of
> > attachment. It only allows N-linking and O-linking glycosylation sites.
> >
> > I would really appreciate if anyone could help give me some
> > advice/suggestions on how to build the structure of levansucrase (1PT2)
> > with a fructosyl residue attached to Asp86. Thank you very much for your
> > help.
> >
> > Regards,
> > Surasak
> >
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
> >
>
>
>
> --
> :-) Lachele
> Lachele Foley
> CCRC/UGA
> Athens, GA USA
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: proposed_mechanism.png)
