Date: Fri, 21 Aug 2015 00:49:14 +0100
Hi,
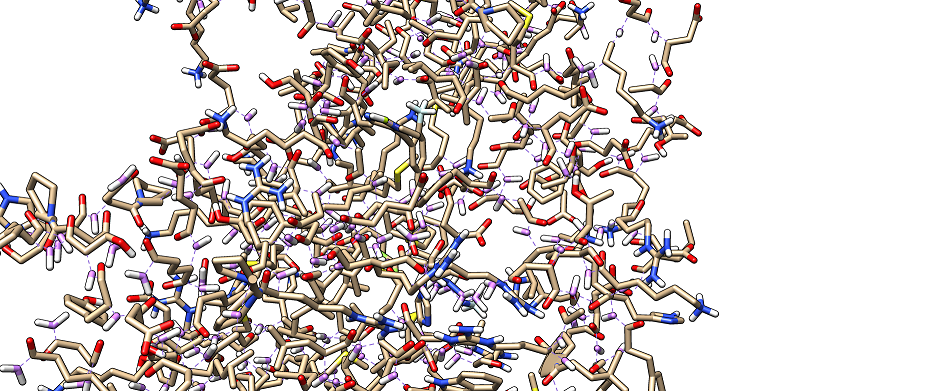
I set up up simulations to use hydrogen mass repartitioning and ran some
simulations with no apparent problems. However, on visualising with
chimera, the protein does not show ribbons which makes visualisation
difficult. Please see attached figure.
The problem only occurs with the topology file created after feeding into
parmed to change masses. I guess the problem is due to atoms being further
apart than the defined distances in chimera? It would be good if somebody
could confirm that this is a known issue and not the result of incorrect
parametrisation? Also, if it is expected behaviour, is there any way to get
ribbon style visualisation back?
many thanks
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: hydrogenMass_chimera-50_.png)
