Date: Thu, 13 Aug 2015 14:58:47 +0800
Hi all£º
I have a question about the method of pH-REMD. I¡¯ve read the paper
published in JCTC(J. Chem. Theory Comput., 2014, 10 (3), pp 1341¨C1352).
It said that the probability of accepting these replica exchange attempts,
given by eq 3, depends only on the difference in the number of titrating
protons present in each replica and their respective difference in pH
I think that it contains an assumption that the residue in lower pH can be
more protonated. But I think that pka of the residue depends the environment
where the residue exists in the protein. And when the proteins structure has
changed severely in lower pH , the residue may have another pH in that
situation. And this situation may contradict with the assumption the theory
contains.
What do you think and I hope to get your explanation.
Thank you!
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
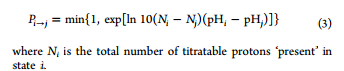
(image/png attachment: image001.png)
