Date: Thu, 30 Apr 2015 17:36:44 +0530
Dear Amber users,
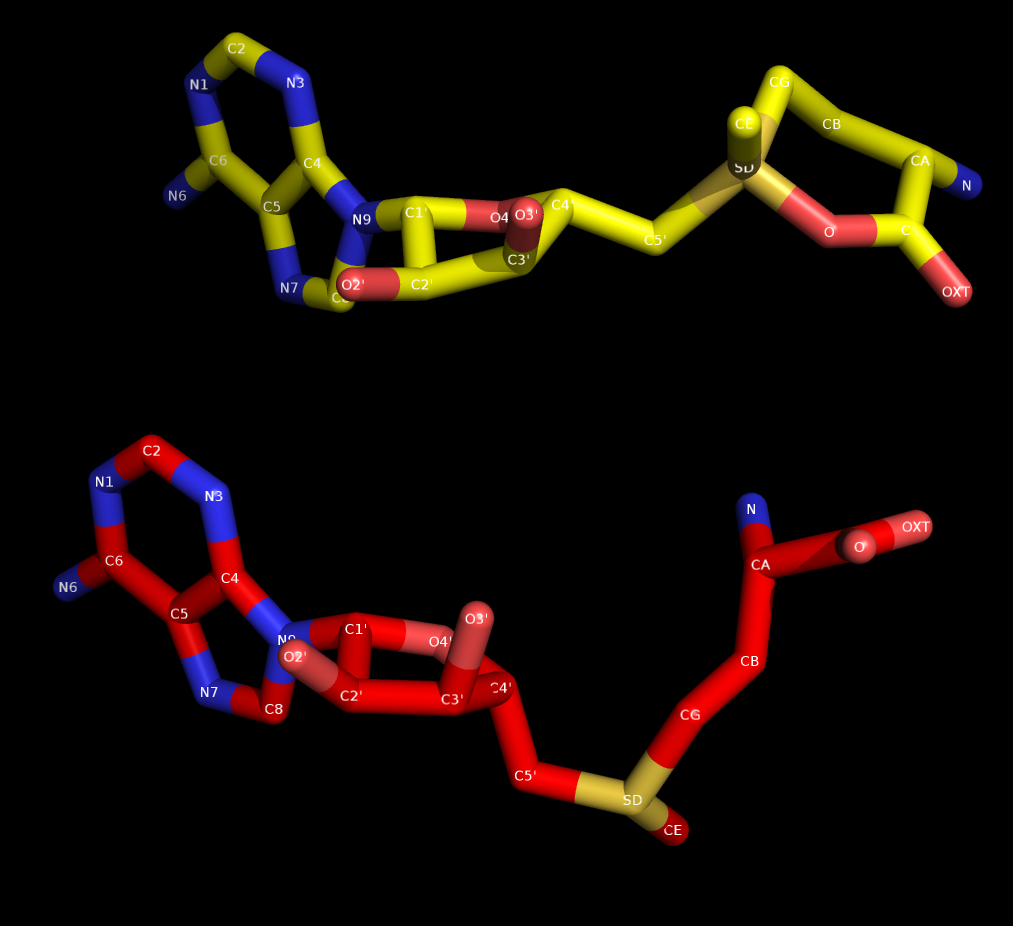
After minimising and heating a protein-ligand complex, I found that the
organic molecule in the protein has an unfavorable conformation that
doesn't match with that of initial structure of the organic molecule (i.e.
before heating), although the protein structure is intact. The pdb file
used for creating frcmod and mol2 is sqm generated, generated after feeding
the pdb file to the antechamber. I have attached a picture showing the
comparison of structures (the yellow colored one is the heated and the red
colored one is the initial structure) along with heat.out, frcmod and mol2
file. Please explain me what is the causation of this structural change and
way to rectify it.
Thanking you.
Swithin Hanosh
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: SAM_comparison.png)
- chemical/x-mol2 attachment: sam1.mol2
- application/octet-stream attachment: sam1.frcmod
- application/octet-stream attachment: 2heat.out
