Date: Tue, 25 May 2010 10:58:53 +0800
Dear All,
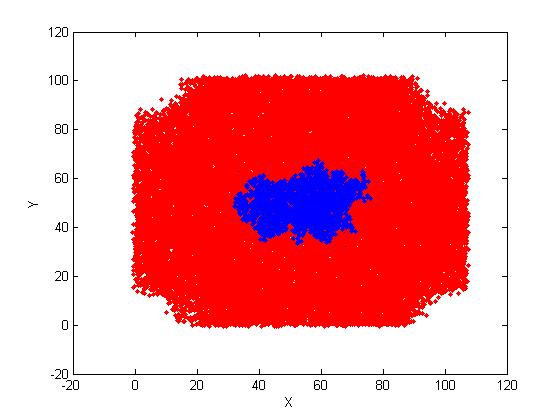
I ran a MD of a case soaking a protein molecule with Sander in a rectangle
PBC box,
but finally the result on file *.rst, are totally with not a rectangle
box. The are 4 missing parts
at the 4 corners of the projection of all the atoms in the the X,Y
directions.
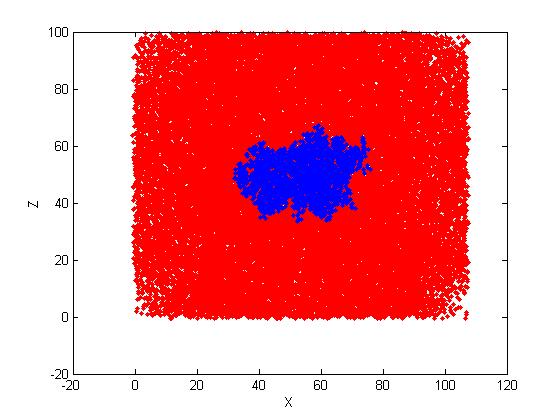
In the direction of X, Z, the projection is still with rectangle shape. Pls
see the attached
projected images. (Water atoms are red; protein atoms are blue).
I don't think it is because of the PBC images because all the atoms look
still to be within the area of this rectangle box and my protein is at the
center of the box.
Can anyone help explain this strange solvation box shape?
The following is the details:
----------------------------------------
I ran a simple case of soaking a protein molecule in a rectangle box:
solvatebox LYZ TIP3PBOX 27
My system is with big dimensions. In the file
of *.inpcrd, the bottom line states:
107.0781340 101.4056200 99.1742170 90.0000000 90.0000000 90.0000000
I ran MD with Sander
0-300K constant temp MD
&cntrl
imin=0,
ntb=1,cut=18.0,
ntc=2, ntf=2,
tempi=0.0, temp0=300.0,
ntt=1,iwrap=1,
nstlim=10000, dt=0.002,
ntpr=500, ntwx=1000
/
Thanks for your help!
Tom
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: Project_XY.jpg)
(image/jpeg attachment: Project_XZ.jpg)
