Date: Sun, 10 Jan 2010 15:34:24 -0500
Dear amber community:
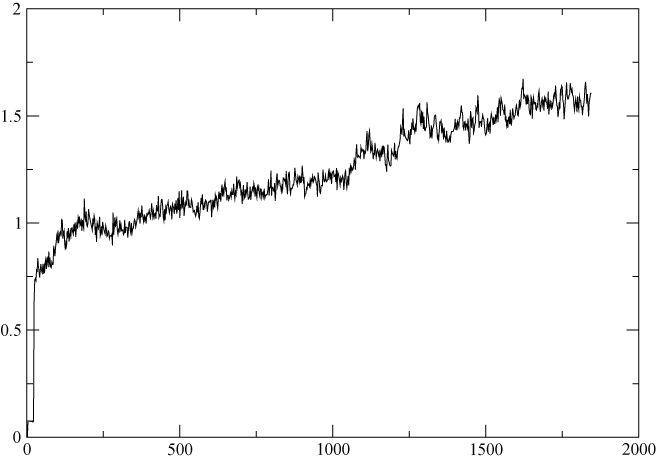
I have run a 20ns simulation for an octomer protein. The trajectory seems OK after visually check in VMD. However, the RMSD vs time plot shows an continuous increased pattern(see attachment) although the range is small, (from 1 to 1.5 Angstrom). It seems to me that a plateau should be reached after equilibration. on the other hand, I think 20ns is long enough for the system to reach equilibrium. here is my question:
Is this an indication of somethin wrong with the simulation? what might be the reason? IF it is normal, What kind of analysis could be used to figure out the origin of RMSD increase?
Your kind of help will be very appreciated. Let me know if further information will help.
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: rmsd.png)
