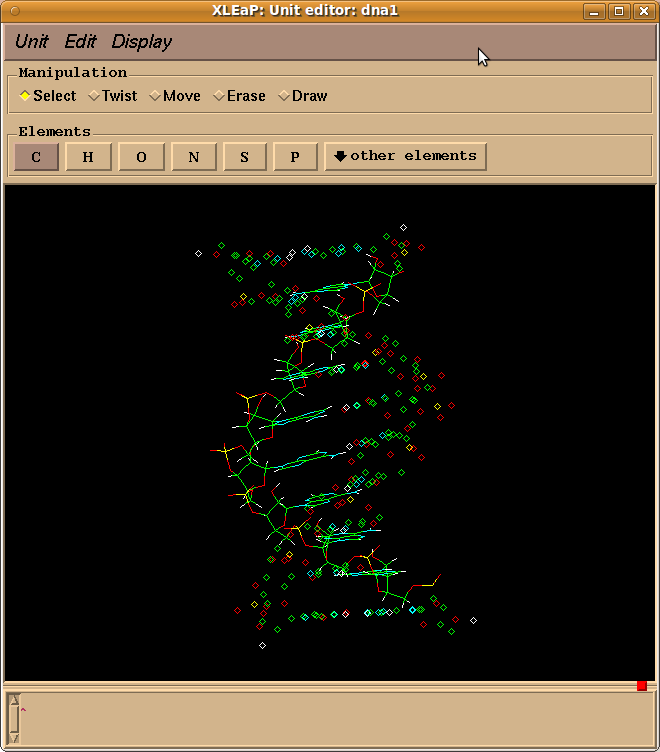
From: Donald Thomas <dthomas21.gmail.com>
Date: Wed, 6 Jan 2010 15:31:02 +1100
Hi,
I'm trying to run through the basic tutorial for building DNA. I have built
Amber 10 on two Ubuntu 9.10 boxes and applied all bugfixes. The
installations were both successful and the tests only result in a handful of
failures that were not significant.
Whether I build the pdb file myself or use the one supplied in the tutorial
I end up with half the DNA not being recognized (even after addition of TER
cards). In xleap it shows up as the attached picture.
It tried AmberTools 1.3 with the same result.
However, if I change to a forcefield other than ff99* such as ff02pol.r1 or
ff03 the file loads perfectly.
Any ideas.
Don Thomas
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
Received on Tue Jan 05 2010 - 21:00:02 PST