Date: Wed, 10 Jun 2009 20:47:58 +0100
Hi all,
I would like just add some additional info regarding
my original question (please read below).
First of all here are precise numbers including standard deviations.
I am also attaching RMSD inormation from both stand alone simulations
and from complex simulation. The red line pointed the moment where
I started with energetical analysis. It seems that some prolongation
of the simulation would be not a bad idea (I have already started with
this)
but I am afraid if the analysis from some extended part of the simulation
affect significantly below reported mean values of dG.
From the attached picture "B_malt_ssDNA_11ns.png" where you can see
discussed complex after 11 ns of simulation is pretty clear that
final binding energy should be here favourable (there is evident nice
complexation) it means negative.
I am also attaching direct mm_pbsa results from both analysis (ONE and
THREE trajectories).
So again, any hints are highly welcomed !
With the best regards
Marek
ONE TRAJECTORY APPROACH
dH - mean value -53,67
dH - standard deviation 13,11
TdS - mean value -74,53
TdS - standard deviation 9,05
dG = dH - TdS - mean value 20,86
dG = dH - TdS - standard deviation 15,93030445
TWO TRAJECTORIES APPROACH
Three trajectory approach
dH - mean value -13,08
dH - standard deviation 57,34
TdS - mean value -79,41
TdS - standard deviation 9,26
dG = dH - TdS - mean value 66,33
dG = dH - TdS - standard deviation 58,08289938
-----------------------MY PREVIOUS
EMAIL------------------------------------------------------------
Dne Tue, 09 Jun 2009 12:49:43 +0200 Marek Maly <marek.maly.ujep.cz>
napsal/-a:
> Dear amber users,
>
> I have done simulation of complex PPI dendrimer - 4th generation which is
> decorated with maltose (whole this molecule is electrically neutral)
> with single strand DNA (25 bases, with charge -24) in salt explicit water
> (ionic strength - 0.15M)- please see the illustrative picture.
>
> The total length of the simulation was 11 ns (4 ns NPT + 7 ns NVT,
> T=310K). Below you can find all "in" files.
> For the parametrisation I used these forcefields - ff99SB (DNA), gaff
> (PPI), glycam (maltose).
>
> Than I made mm_PBSA energetical analysis using the last 2.5 ns (I took 50
> frames in 0.05 ns interval).
>
> Here are the final results:
>
> One trajectory approach:
>
> dG = dH - TdS = -53.67 - (-74.53) = 20.86
>
> Three trajectories approach:
>
> dG = dH - TdS = -15.27 - (-78.67) = 63.4
>
> Maybe it is worth to notice that to speed up the NN analysis I used
> separate parallel sander minimisation (conj-grad) using input file
> generated
> by mm_pbsa routine:
>
> -----------------------------------------------
> File generated by mm_pbsa.pl
> &cntrl
> ntxo = 1,
> ntf = 1, ntb = 0,
> dielc = 4,
> cut = 99.0, nsnb = 99999,
> scnb = 2.0, scee = 1.2,
> imin = 1, maxcyc = 100000,
> ncyc = 0, drms = 0.0001,
> &end
> &ewald
> eedmeth= 5,
> &end
> ----------------------------------------------
>
>
> I am really not sure how to interpret above reported results, I am not
> sure if they are even reliable ...
>
> I would be really grateful for any comments to this strange results.
>
> It is worth to play with some mm_pbsa parameters (to obtain lower dH
> contribution and so less positive dG ?)
> or should I try to repeat all the analysis with NAB instead of mm_PBSA
> routine ?
>
> Thank you very much in advance for any ideas !
>
> With the best regards
>
> Marek
>
>
>
>
>
> Please find below all the important files/data:
>
>
> ///////THE INPUT FILES FOR MINIMISATION AND
> MD////////////////////////////////
> #1 - minimisation with restraints
>
> minimise ras-raf
> &cntrl
> imin=1,
> ntmin = 1,
> maxcyc=5000,
> ncyc=500,
> cut=10.0,
> ntb=1,
> ntpr=100,
> ntr=1,
> restraintmask=':1-150',
> restraint_wt=5.0,
> /
> #2 - minimistaion without restraints
>
> minimise ras-raf
> &cntrl
> imin=1,maxcyc=5000,ncyc=2000,
> cut=10.0,ntb=1,
> ntpr=100,
> /
> #3 - heating
>
> heat ras-raf
> &cntrl
> imin=0,irest=1,ntx=5,
> nstlim=50000,dt=0.001,
> ntc=2,ntf=2,
> cut=10.0, ntb=2,
> ntp=1,
> taup=1.0,
> ntpr=5000, ntwx=5000,
> ntt=3, gamma_ln=2.0,
> temp0=310.0,
> ig=-1,
> ntr=1, restraintmask=':1-150',
> restraint_wt=2.0,
> /
> #4 - density
> heat ras-raf
> &cntrl
> imin=0,irest=1,ntx=5,
> nstlim=250000,dt=0.002,
> ntc=2,ntf=2,
> cut=10.0, ntb=2, ntp=1, taup=2.0,
> ntpr=50000, ntwx=50000,
> ntt=3, gamma_ln=2.0,
> ig=-1,
> temp0=310.0,
> /
> #5 - 4 ns NPT (equilibrium)
> heat ras-raf
> &cntrl
> imin=0,irest=1,ntx=5,
> nstlim=250000,dt=0.002,
> ntc=2,ntf=2,
> cut=10.0, ntb=2, ntp=1, taup=2.0,
> ntpr=50000, ntwx=50000,
> ntt=3, gamma_ln=2.0,
> ig=-1,
> temp0=310.0,
> /
> #6 - 7 ns NVT (production)
>
> heat ras-raf
> &cntrl
> imin=0,irest=1,ntx=5,
> nstlim=250000,dt=0.002,
> ntc=2,ntf=2,
> cut=10.0, ntb=1,
> ntpr=5000, ntwx=5000,
> ntt=3, gamma_ln=2.0,
> ig=-1,
> temp0=310.0,
> /
>
>
> /////AVERAGES from the last 0.5 ns of simulation
>
>>>> COMPLEX
>
> A V E R A G E S O V E R 50 S T E P S
>
>
> NSTEP = 250000 TIME(PS) = 11100.000 TEMP(K) = 310.11 PRESS
> = 0.0
> Etot = -156998.5026 EKtot = 39280.1612 EPtot =
> -196278.6638
> BOND = 2250.7410 ANGLE = 1813.0047 DIHED =
> 1060.0015
> 1-4 NB = 639.7923 1-4 EEL = 7345.4528 VDWAALS =
> 27593.2769
> EELEC = -236980.9330 EHBOND = 0.0000 RESTRAINT =
> 0.0000
> Ewald error estimate: 0.4593E-04
>
>
>>>> RECEPTOR(dendrimer) - seaparate run
>
> ------------------------------------------------------------------------------
>
>
> A V E R A G E S O V E R 50 S T E P S
>
>
> NSTEP = 250000 TIME(PS) = 11100.000 TEMP(K) = 309.89 PRESS
> = 0.0
> Etot = -116491.9660 EKtot = 30052.9725 EPtot =
> -146544.9385
> BOND = 2031.8582 ANGLE = 1382.5666 DIHED =
> 496.8121
> 1-4 NB = 440.5337 1-4 EEL = 10396.5931 VDWAALS =
> 21710.7831
> EELEC = -183004.0852 EHBOND = 0.0000 RESTRAINT =
> 0.0000
> Ewald error estimate: 0.5800E-04
>
>
>
>>>> LIGAND(DNA) - separate run
>
> A V E R A G E S O V E R 50 S T E P S
>
>
> NSTEP = 250000 TIME(PS) = 11100.000 TEMP(K) = 309.94 PRESS
> = 0.0
> Etot = -91503.2207 EKtot = 20857.8449 EPtot =
> -112361.0656
> BOND = 206.6247 ANGLE = 447.1949 DIHED =
> 527.7110
> 1-4 NB = 213.5335 1-4 EEL = -3008.4689 VDWAALS =
> 14905.0176
> EELEC = -125652.6782 EHBOND = 0.0000 RESTRAINT =
> 0.0000
> Ewald error estimate: 0.6680E-04
>
>
>
>
>
>
> //////ENERGETICAL ANALYSIS using mm_PBSA (reported are just files for
> single trajectory approach/ in the case
> of 3 traj I used the same just with minor changes in )
>
>
>
> *******ENTHALPY CONTRIBUTION
> *******ENTHALPY CONTRIBUTION
> *******ENTHALPY CONTRIBUTION
> *******ENTHALPY CONTRIBUTION
>
> #
> # Input parameters for mm_pbsa.pl
> #
> # Holger Gohlke
> # 08.01.2002
> #
> ################################################################################
> .GENERAL
> #
> # General parameters
> # 0: means NO; >0: means YES
> #
> # mm_pbsa allows to calculate (absolute) free energies for one
> molecular
> # species or a free energy difference according to:
> #
> # Receptor + Ligand = Complex,
> # DeltaG = G(Complex) - G(Receptor) - G(Ligand).
> #
> # PREFIX - To the prefix, "{_com, _rec, _lig}.crd.Number" is added
> during
> # generation of snapshots as well as during mm_pbsa
> calculations.
> # PATH - Specifies the location where to store or get snapshots.
> #
> # COMPLEX - Set to 1 if free energy difference is calculated.
> # RECEPTOR - Set to 1 if either (absolute) free energy or free energy
> # difference are calculated.
> # LIGAND - Set to 1 if free energy difference is calculated.
> #
> # COMPT - parmtop file for the complex (not necessary for option GC).
> # RECPT - parmtop file for the receptor (not necessary for option GC).
> # LIGPT - parmtop file for the ligand (not necessary for option GC).
> #
> # GC - Snapshots are generated from trajectories (see below).
> # AS - Residues are mutated during generation of snapshots from
> trajectories.
> # DC - Decompose the free energies into individual contributions
> # (only works with MM and GB).
> #
> # MM - Calculation of gas phase energies using sander.
> # GB - Calculation of desolvation free energies using the GB models in
> sander
> # (see below).
> # PB - Calculation of desolvation free energies using delphi (see
> below).
> # Calculation of nonpolar solvation free energies according to
> # the NPOPT option in pbsa (see below).
> # MS - Calculation of nonpolar contributions to desolvation using
> molsurf
> # (see below).
> # If MS == 0 and GB == 1, nonpolar contributions are calculated
> with the
> # LCPO method in sander.
> # If MS == 0 and PB == 1, nonpolar contributions are calculated
> according
> # the NPOPT option in pbsa (see below).
> # NM - Calculation of entropies with nmode.
> #
> PREFIX snap
> PATH ./
> #
> COMPLEX 1
> RECEPTOR 1
> LIGAND 1
> #
> COMPT ./com.prmtop
> RECPT ./rec.prmtop
> LIGPT ./lig.prmtop
> #
> GC 0
> AS 0
> DC 0
> #
> MM 1
> GB 0
> PB 1
> MS 1
> #
> NM 0
> #
> ################################################################################
> .PB
> #
> # PB parameters (this section is only relevant if PB = 1 above)
> #
> # The following parameters are passed to the PB solver.
> # Additional input parameters may also be added here. See the sander PB
> # documentation for more options.
> #
> # PROC - Determines which method is used for solving the PB equation:
> # By default, PROC = 2, the pbsa program of the AMBER suite is
> used.
> # REFE - Determines which reference state is taken for PB calc:
> # By default, REFE = 0, reaction field energy is calculated
> with
> # EXDI/INDI. Here, INDI must agree with DIELC from MM part.
> # INDI - Dielectric constant for the solute.
> # EXDI - Dielectric constant for the surrounding solvent.
> # ISTRNG - Ionic strength (in mM) for the Poisson-Boltzmann solvent.
> # PRBRAD - Solvent probe radius in Angstrom:
> # 1.4: with the radii in the prmtop files. Default.
> # 1.6: with the radii optimized by Tan and Luo (In
> preparation).
> # See RADIOPT on how to choose a cavity radii set.
> # RADIOPT - Option to set up radii for PB calc:
> # 0: uses the radii from the prmtop file. Default.
> # 1: uses the radii optimized by Tan and Luo (In preparation)
> # with respect to the reaction field energies computed
> # in the TIP3P explicit solvents. Note that optimized radii
> # are based on AMBER atom types (upper case) and charges.
> # Radii from the prmtop files are used if the atom types
> # are defined by antechamber (lower case).
> # SCALE - Lattice spacing in no. of grids per Angstrom.
> # LINIT - No. of iterations with linear PB equation.
> #
> # NP Parameters for nonpolar solvation energies if MS = 0
> #
> # NPOPT - Option for modeling nonpolar solvation free energy.
> # See sander PB documentation for more information on the
> # implementations by Tan and Luo (In preparation).
> # 1: uses the solvent-accessible-surface area to correlate
> total
> # nonpolar solvation free energy:
> # Gnp = CAVITY_SURFTEN * SASA + CAVITY_OFFSET. Default.
> # 2: uses the solvent-accessible-surface area to correlate the
> # repulsive (cavity) term only, and uses a surface-integration
> # approach to compute the attractive (dispersion) term:
> # Gnp = Gdisp + Gcavity
> # = Gdisp + CAVITY_SURFTEN * SASA + CAVITY_OFFSET.
> # When this option is used, RADIOPT has to be set to 1,
> # i.e. the radii set optimized by Tan and Luo to mimic Gnp
> # in TIP3P explicit solvents. Otherwise, there is no guarantee
> # that Gnp matches that in explicit solvents.
> # CAVITY_SURFTEN/CAVITY_OFFSET - Values used to compute the nonpolar
> # solvation free energy Gnp according NPOPT. The default values
> # are for NPOPT set to 0 and RADIOPT set to 0 (see above).
> # If NPOPT is set to 1 and RADIOPT set to 1, these two lines
> # can be removed, i.e. use the default values set in pbsa
> # for this nonpolar solvation model. Otherwise, please
> # set these to the following:
> # CAVITY_SURFTEN: 0.04356
> # CAVITY_OFFSET: -1.008
> #
> # NP Parameters for nonpolar solvation energies if MS = 1
> #
> # SURFTEN/SURFOFF - Values used to compute the nonpolar contribution
> Gnp
> to
> # the desolvation according to Gnp = SURFTEN * SASA + SURFOFF.
> #
> PROC 2
> REFE 0
> INDI 1.0
> EXDI 80.0
> SCALE 2
> LINIT 1000
> PRBRAD 1.4
> ISTRNG 0.0
> RADIOPT 0
> NPOPT 1
> CAVITY_SURFTEN 0.0072
> CAVITY_OFFSET 0.00
> #
> SURFTEN 0.0072
> SURFOFF 0.00
> #
> ################################################################################
> .MM
> #
> # MM parameters (this section is only relevant if MM = 1 above)
> #
> # The following parameters are passed to sander.
> # For further details see the sander documentation.
> #
> # DIELC - Dielectricity constant for electrostatic interactions.
> # Note: This is not related to GB calculations.
> #
> DIELC 1.0
> #
> ################################################################################
> .GB
> #
> # GB parameters (this section is only relevant if GB = 1 above)
> #
> # The first group of the following parameters are passed to sander.
> # For further details see the sander documentation.
> #
> # IGB - Switches between Tsui's GB (1), Onufriev's GB (2, 5).
> # GBSA - Switches between LCPO (1) and ICOSA (2) method for SASA calc.
> # Decomposition only works with ICOSA.
> # SALTCON - Concentration (in M) of 1-1 mobile counterions in solution.
> # EXTDIEL - Dielectricity constant for the solvent.
> # INTDIEL - Dielectricity constant for the solute
> #
> # SURFTEN / SURFOFF - Values used to compute the nonpolar contribution
> Gnp to
> # the desolvation according to Gnp = SURFTEN * SASA +
> SURFOFF.
> #
> IGB 2
> GBSA 1
> SALTCON 0.00
> EXTDIEL 80.0
> INTDIEL 1.0
> #
> SURFTEN 0.0072
> SURFOFF 0.00
> #
> ################################################################################
> .MS
> #
> # Molsurf parameters (this section is only relevant if MS = 1 above)
> #
> # PROBE - Radius of the probe sphere used to calculate the SAS.
> # Since Bondi radii are already augmented by 1.4A, PROBE should
> be 0.0
> #
> PROBE 0.0
> #
> #################################################################################
> .PROGRAMS
> #
> # Additional program executables can be defined here
> #
> #
> ################################################################################
>
>
>
>
> ************INPUT FILE for NN analysis
> ************INPUT FILE for NN analysis
> ************INPUT FILE for NN analysis
> ************INPUT FILE for NN analysis
>
> #
> # Input parameters for mm_pbsa.pl
> #
> # Holger Gohlke
> # 08.01.2002
> #
> ################################################################################
> .GENERAL
> #
> # General parameters
> # 0: means NO; >0: means YES
> #
> # mm_pbsa allows to calculate (absolute) free energies for one
> molecular
> # species or a free energy difference according to:
> #
> # Receptor + Ligand = Complex,
> # DeltaG = G(Complex) - G(Receptor) - G(Ligand).
> #
> # PREFIX - To the prefix, "{_com, _rec, _lig}.crd.Number" is added
> during
> # generation of snapshots as well as during mm_pbsa
> calculations.
> # PATH - Specifies the location where to store or get snapshots.
> #
> # COMPLEX - Set to 1 if free energy difference is calculated.
> # RECEPTOR - Set to 1 if either (absolute) free energy or free energy
> # difference are calculated.
> # LIGAND - Set to 1 if free energy difference is calculated.
> #
> # COMPT - parmtop file for the complex (not necessary for option GC).
> # RECPT - parmtop file for the receptor (not necessary for option GC).
> # LIGPT - parmtop file for the ligand (not necessary for option GC).
> #
> # GC - Snapshots are generated from trajectories (see below).
> # AS - Residues are mutated during generation of snapshots from
> trajectories.
> # DC - Decompose the free energies into individual contributions
> # (only works with MM and GB).
> #
> # MM - Calculation of gas phase energies using sander.
> # GB - Calculation of desolvation free energies using the GB models in
> sander
> # (see below).
> # PB - Calculation of desolvation free energies using delphi (see
> below).
> # Calculation of nonpolar solvation free energies according to
> # the NPOPT option in pbsa (see below).
> # MS - Calculation of nonpolar contributions to desolvation using
> molsurf
> # (see below).
> # If MS == 0 and GB == 1, nonpolar contributions are calculated
> with the
> # LCPO method in sander.
> # If MS == 0 and PB == 1, nonpolar contributions are calculated
> according
> # the NPOPT option in pbsa (see below).
> # NM - Calculation of entropies with nmode.
> #
> PREFIX snap
> PATH ./
> #
> COMPLEX 1
> RECEPTOR 1
> LIGAND 1
> #
> COMPT ./com.prmtop
> RECPT ./rec.prmtop
> LIGPT ./lig.prmtop
> #
> GC 0
> AS 0
> DC 0
> #
> MM 0
> GB 0
> PB 0
> MS 0
> #
> NM 1
> #
> #
> #################################################################################
> .NM
> #
> # Parameters for sander/nmode calculation (this section is only relevant
> # if NM = 1 above)
> #
> # The following parameters are passed to sander (for minimization) and
> nmode
> # (for entropy calculation using gasphase statistical mechanics).
> # For further details see documentation.
> #
> # DIELC - (Distance-dependent) dielectric constant
> # MAXCYC - Maximum number of cycles of minimization.
> # DRMS - Convergence criterion for the energy gradient.
> #
> DIELC 4
> MAXCYC 30
> DRMS 0.0001
> #
> #################################################################################.PROGRAMS
> #
> # Additional program executables can be defined here
> #
> #
> ################################################################################
>
>
>
>
>
>
>
>
>
-- Tato zpráva byla vytvořena převratným poštovním klientem Opery: http://www.opera.com/mail/
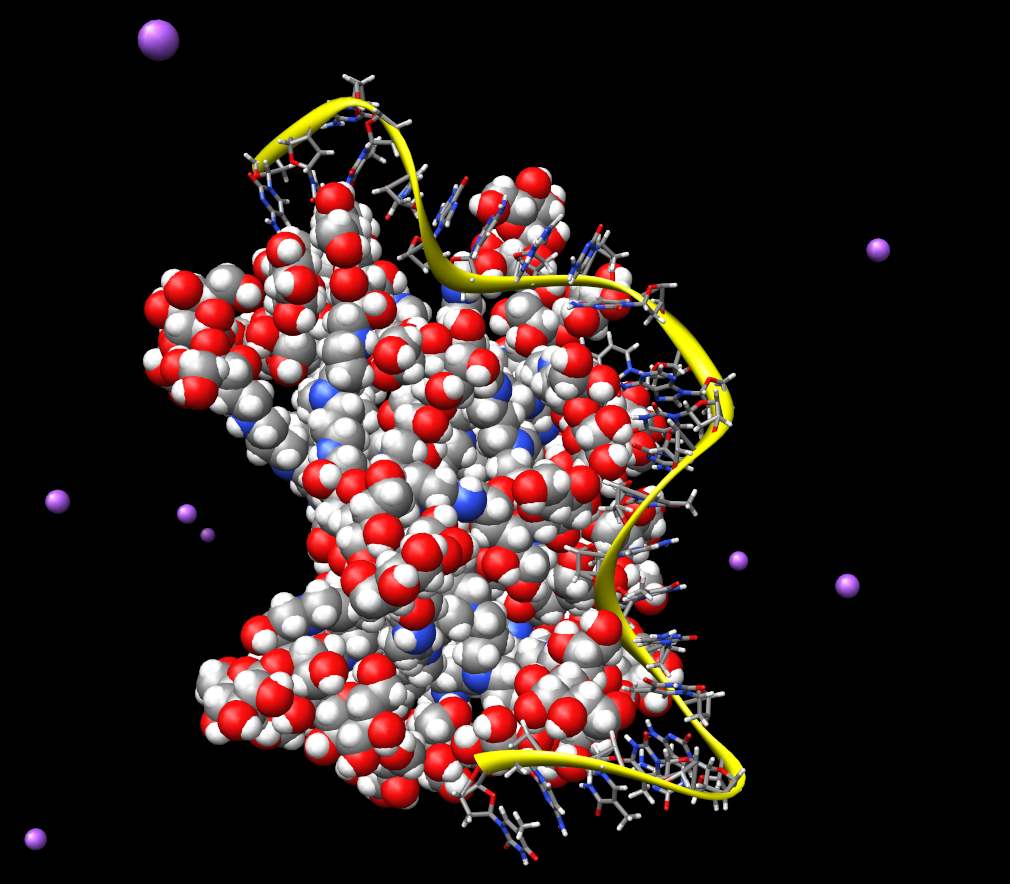
(image/png attachment: B_malt_ssDNA_11ns.png)
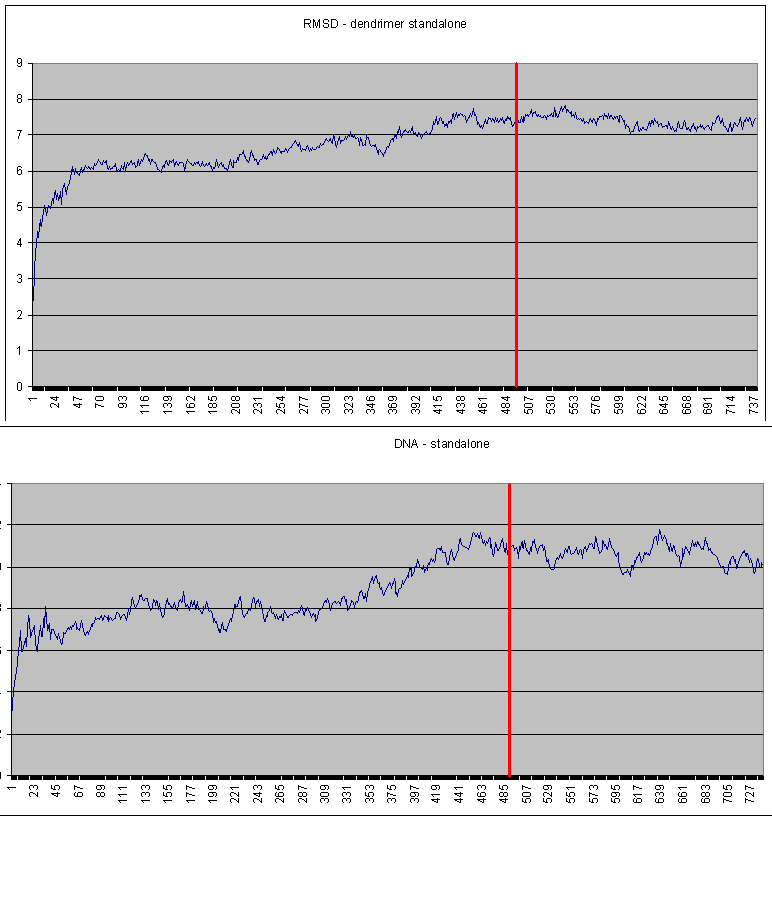
(image/png attachment: RMSD-standalone.png)
(image/png attachment: RMSD-COMPLEX.PNG)
- application/octet-stream attachment: snap_statistics-THREE-TRAJ-APPROACH.out
- application/octet-stream attachment: snap_statistics-ONE-TRAJ-APPROACH.out
- text/plain attachment: ATT00002.txt
