Date: Thu, 4 Sep 2025 23:23:22 +0000
Hi Ge,
I have good news and bad news.
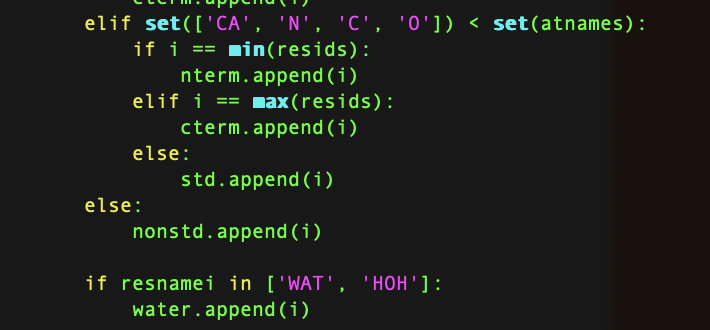
Good news first, this error is due to PyMST mistakenly considering SAM as the C-terminal. You may find the source code snippet here at "amber/lib/python3.12/site-packages/pymsmt/mol/mol.py"
[cid:fe435964-f5ad-45b4-a7b4-58d4bda7e7f0]
I other words, PyMSMT will consider the last residue that has any atoms from the list "CA, N, C, O" as the C-terminal residue. And luckily, you have those in the SAM molecule.
Please see the "1TV8_modified_a_little.zip" folder that I have DM-ed you. This folder contains my fixes on this issue by simply renaming the atoms in SAM, both in PDB and MOL2.
Then comes the bad news: this 1TV8 system is extremely hard to prepare using MCPB.py, mostly because it has multiple metal centers with different charges. Please see the "1TV8_modified_a_lot.zip" folder that I have DM-ed you. Here, I named each iron uniquely by calling them FE6, FE7, FE8 and FE9 (I forgot to change FE back to FE6 in the second row of FE6.mol2 after a test).
Only by this method will the "chargedict" in "gauio.py" register charges separately; otherwise, all FE will have the same charge. But if they are named uniquely, the next step in "gauio.py" will cry out, "Could not find VDW radius for element FE6" because it uses a different ion dictionary (called "IonLJParaDict" in amber/lib/python3.12/site-packages/pymsmt/mol/element.py) to build the VDW terms for Gaussian input files, and there is no FE6, FE7, FE8, or FE9 in that dictionary.
I can manually add those ions to the dictionary, but no guarantee that it will work for every subsequent step.
Best regards,
Zhen.
_____________________
Zhen Li<http://lizhen62017.wixsite.com/home>, Ph.D.,
The Merz Research Group<http://merzgroup.org>,
Michigan State University,
Cleveland Clinic.
________________________________
From: Ge Song <g.song.duke.edu>
Sent: Thursday, September 4, 2025 3:09 PM
To: Li, Zhen <lizhen6.chemistry.msu.edu>; amber <amber.ambermd.org>
Subject: Re: KeyError of chargedict in MCPB's step 1
Dear Zhen,
Thanks. I have reduce it to include the hydrogens. But I still got the same error with the new mol2 file.
Best,
Ge
________________________________
From: Li, Zhen <lizhen6.chemistry.msu.edu>
Sent: Thursday, September 4, 2025 2:48 PM
To: Ge Song <g.song.duke.edu>; amber <amber.ambermd.org>
Subject: Re: KeyError of chargedict in MCPB's step 1
Hi Ge,
After reviewing the files, several tasks need to be completed before sending them to MCPB.py.
1.
SAM.mol2 is not reduced to include hydrogens. The command "reduce SAM.pdb > SAM_H.pdb" must be run, and then the SAM_H.mol2 file needs to be generated.
2.
After fixing SAM, could you also send me the SF4.mol2 file? I am unable to execute the MCPB.py program without this file.
Best regards,
Zhen.
_____________________
Zhen Li<https://urldefense.com/v3/__http://lizhen62017.wixsite.com/home__;!!HXCxUKc!wOgLC1p0-OK7ENK6j1JJxWyxsJLCvS-RB5d1P_uOGANqrqZZFneB-xMVYiCmnBNenPU5W8W19vS4wzi27Pl67yDn$>, Ph.D.,
The Merz Research Group<https://urldefense.com/v3/__http://merzgroup.org/__;!!HXCxUKc!wOgLC1p0-OK7ENK6j1JJxWyxsJLCvS-RB5d1P_uOGANqrqZZFneB-xMVYiCmnBNenPU5W8W19vS4wzi27ORD0VIw$>,
Michigan State University,
Cleveland Clinic.
________________________________
From: Ge Song <g.song.duke.edu>
Sent: Thursday, September 4, 2025 1:18 PM
To: Li, Zhen <lizhen6.chemistry.msu.edu>; amber <amber.ambermd.org>
Subject: Re: KeyError of chargedict in MCPB's step 1
Dear Zhen,
Thanks for your instrcutions. But my mol2 file is generated by antechamber and has a unique name for each atom.
I have shared the files I am using to your email. I really appreciate your time!
Best regards,
Ge
________________________________
From: Li, Zhen <lizhen6.chemistry.msu.edu>
Sent: Thursday, September 4, 2025 12:55 PM
To: amber <amber.ambermd.org>; Ge Song <g.song.duke.edu>
Subject: Re: KeyError of chargedict in MCPB's step 1
Hi Ge,
Thank you for trying MCPB.py.
Unfortunately, I am unable to get the files that you shared. It states that the files are only viewable using a SharePoint account registered with "amber.ambermd.org", but I don't think the AMBER mailing list account is associated with SharePoint. Could you please share those with liz7.ccf.org? Or simply attach part of the PDB and MOL2 text in the email? Appreciate your efforts.
[cid:fc68c507-21ea-4a7a-99d0-714361b4bb31]
With current clues, I guess that your SAM residue has several "C"s and "H"s of the same name, and so on. Pengfei's 06/11/2020 email states that not only shall the atom name match between PDB and MOL2, but the atom name within a residue should also be unique.
I understand that it is a "pain in the neck" to give each atom its unique name by hand for SAM (which has about 50 atoms...). But programs like antechamber will help. Simply run "antechamber -i <input_pdb> -fi pdb -o <output_pdb> -fo pdb", and it will rename the atoms for you.
Looking forward to more updates.
Best regards,
Zhen
_____________________
Zhen Li<https://urldefense.com/v3/__http://lizhen62017.wixsite.com/home__;!!HXCxUKc!3-tqHi9Nr8XCm9sTsGlMd3mYAMs3hx_csvouJ69mAhQDKY--g106HtD1KtR_7IiBu4yQwoeHzUqISiktrW80TZbA$>, Ph.D.,
The Merz Research Group<https://urldefense.com/v3/__http://merzgroup.org/__;!!HXCxUKc!3-tqHi9Nr8XCm9sTsGlMd3mYAMs3hx_csvouJ69mAhQDKY--g106HtD1KtR_7IiBu4yQwoeHzUqISiktrWyHMtVB$>,
Michigan State University,
Cleveland Clinic.
________________________________
From: Ge Song via AMBER <amber.ambermd.org>
Sent: Thursday, September 4, 2025 11:47 AM
To: Pengfei Li via AMBER <amber.ambermd.org>
Subject: [AMBER] KeyError of chargedict in MCPB's step 1
Dear Amber developers,
I have got the same error in running MCPB step 1 as posted here before (https://urldefense.com/v3/__http://archive.ambermd.org/202006/0052.html__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvHkfRfRo$ ):
File "/home/software/AMBER/19/amber18/lib/python2.7/site-packages/pymsmt/mcpb/gene_model_files.py", line 1690, in gene_model_files
totchg = totchg + chargedict['C' + mol.residues[i].resname]
KeyError: 'CSAM'
According to the previous reply from pengfei (https://urldefense.com/v3/__http://archive.ambermd.org/202006/0100.html__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIv3_SmQyc$ ), matching the atom names in mol2 file with original_pdb file should solve the problem, but this is not the case for me.
I did include the SAM.mol2 file:
original_pdb 1TV8_cym.pdb
group_name fs_clusters
ion_ids 5308 5309 5310 5311 5316 5317 5318 5319
ion_mol2files FE2.mol2 FE2.mol2 FE2.mol2 FE3.mol2 FE2.mol2 FE2.mol2 FE2.mol2 FE2.mol2
naa_mol2files SF4.mol2 SAM.mol2 SO4.mol2
force_field ff14SB
water_model tip3p
cut_off 2.8
charge_method resp
qm_theory B3LYP
basis_set 6-31G*
And the atom names exactly match with the original pdb.
I am wondering if anyone could give me some suggestions (I have attached the 1TV8_cym.pdb abd all mol2 files I am using).
Best,
Ge
[https://urldefense.com/v3/__https://res.public.onecdn.static.microsoft/assets/mail/file-icon/png/generic_16x16.png*5D1TV8_cym.pdb__;JQ!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvlrTMxDQ$ <https://urldefense.com/v3/__https://prodduke-my.sharepoint.com/:u:/g/personal/gs298_duke_edu/EY-yYHosL01PtggK1cHP1DEBciKErRX0io073Hs5dtVvRw__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvz72O7_w$ >
[https://urldefense.com/v3/__https://res.public.onecdn.static.microsoft/assets/mail/file-icon/png/generic_16x16.png*5DFE2.mol2__;JQ!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvSWhwgu0$ <https://urldefense.com/v3/__https://prodduke-my.sharepoint.com/:u:/g/personal/gs298_duke_edu/EZEKOVjfMplAsp9RJsR71OoB8ffpHpd9qoZ5je2TcMY5pQ__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvZEjvi50$ >
[https://urldefense.com/v3/__https://res.public.onecdn.static.microsoft/assets/mail/file-icon/png/generic_16x16.png*5DFE3.mol2__;JQ!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvUFR2f3M$ <https://urldefense.com/v3/__https://prodduke-my.sharepoint.com/:u:/g/personal/gs298_duke_edu/EevdAaEWv1JComYBrqRcFpsB5PQ4XwLSJArNmO-6EGN1Sw__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvr0avaC8$ >
[https://urldefense.com/v3/__https://res.public.onecdn.static.microsoft/assets/mail/file-icon/png/generic_16x16.png*5DSAM.mol2__;JQ!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvYcStCCI$ <https://urldefense.com/v3/__https://prodduke-my.sharepoint.com/:u:/g/personal/gs298_duke_edu/ESH1S34rxL9OgoJPUfAvWoQBUzPYkYQ1tokE3qruujDvfA__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIviWUuSQg$ >
[https://urldefense.com/v3/__https://res.public.onecdn.static.microsoft/assets/mail/file-icon/png/generic_16x16.png*5DSO4.mol2__;JQ!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIv1J9DOm0$ <https://urldefense.com/v3/__https://prodduke-my.sharepoint.com/:u:/g/personal/gs298_duke_edu/EYVxx8w2LOxBk7n1HLvxObsBaBMVsBU5KQhRQQdi_zeE4w__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIvzJlqq4M$ >
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
https://urldefense.com/v3/__http://lists.ambermd.org/mailman/listinfo/amber__;!!HXCxUKc!3NxkDUfg2DRUN5t8UexsJq2_s9RvndxHde4ApcWEH1AGbdPCITTEP1kiJt5nptFrZjimcHiDF4UAUWIv3IYyauY$
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: 02-image.png)
