Date: Fri, 4 Aug 2023 02:11:18 +0000
Dear Amber list members,
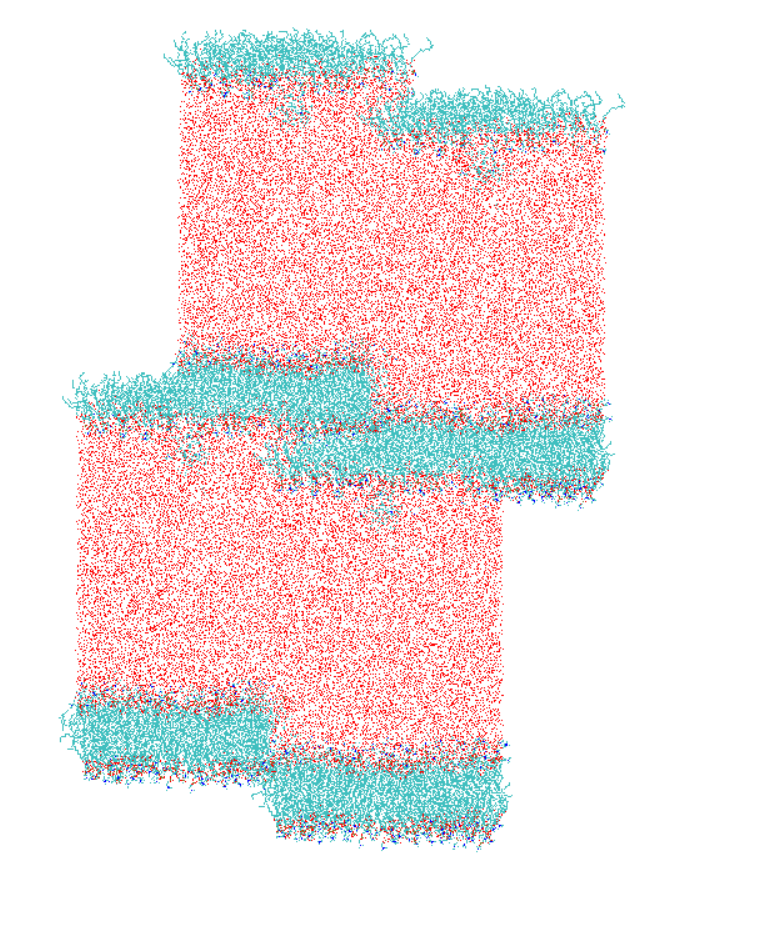
I am simulating a POPC bilayer in water following the default CHARMM-GUI protocol. On visualizing the production trajectory, I saw a weird behavior where the bilayer and water layer positions had been flipped. I know theoretically this doesn't mean anything given the images in periodic boxes, and probably this shouldn't affect the resulting properties. However, I am hoping for a reason for this and a way to produce a nice trajectory with it. I have attached a snapshot from after 10 ns of NPT production. You would see that there is a shift between periodic boxes along the Z axis (trajectory aligned to the bilayer residues). I appreciate your expert thoughts on this.
A NPT simulation for common production-level simulations
&cntrl
imin=0, ! No minimization
irest=1, ! This IS a restart of an old MD simulation
ntx=5, ! So our inpcrd file has velocities
! Temperature control
ntt=3, ! Langevin dynamics
gamma_ln=1.0, ! Friction coefficient (ps^-1)
temp0=310, ! Target temperature
! Potential energy control
cut=9.0, ! nonbonded cutoff, in Angstroms
! MD settings
nstlim=5000000,! 10 ns total
dt=0.002, ! time step (ps)
! SHAKE
ntc=2, ! Constrain bonds containing hydrogen
ntf=2, ! Do not calculate forces of bonds containing hydrogen
! Control how often information is printed
ntpr=20000, ! Print energies every 10000 steps
ntwx=10000, ! Print coordinates every 50000 steps to the trajectory
ntwr=2500000, ! Print a restart file every 2500K steps (can be less frequent)
! ntwv=-1, ! Uncomment to also print velocities to trajectory
! ntwf=-1, ! Uncomment to also print forces to trajectory
ntxo=2, ! Write NetCDF format
ioutfm=1, ! Write NetCDF format (always do this!)
! Wrap coordinates when printing them to the same unit cell
iwrap=1,
! Constant pressure control.
barostat=1, ! Berendsen barostat... change to 2 for MC
ntp=3, ! 1=isotropic, 2=anisotropic, 3=semi-isotropic w/ surften
pres0=1.0, ! Target external pressure, in bar
taup=1.0,
! Constant surface tension (needed for semi-isotropic scaling). Uncomment
! for this feature. csurften must be nonzero if ntp=3 above
csurften=3, ! Interfaces in 1=yz plane, 2=xz plane, 3=xy plane
gamma_ten=0.0, ! Surface tension (dyne/cm). 0 gives pure semi-iso scaling
ninterface=2, ! Number of interfaces (2 for bilayer)
! Set water atom/residue names for SETTLE recognition
watnam='WAT', ! Water residues are named WAT
owtnm='O', ! Water oxygens are named O
/
Thank you,
Senal Liyanage
[cid:fe008a5e-0d6d-4c64-b803-b54a918858e8]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
