Date: Mon, 25 Jan 2021 08:37:48 +0000
Dear amber group,
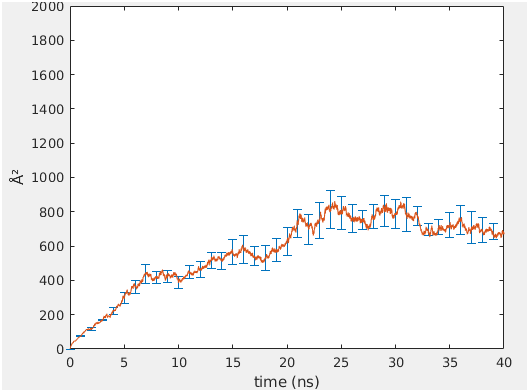
Is there any issue with the diffusion command in cpptraj amber-16? For a 66-lipid micelle we get a plateau value that does not follow a linear relationship and we have not seen this in other systems. There seems not to be a drift in the electrostatic interactions. This is a NVT simulation.
The commands in ptraj are as follows:
trajin Simu1_run1.mdcrd 1 100000 10
trajin Simu1_run2.mdcrd 1 100000 10
trajin Simu1_run3.mdcrd 1 100000 10
trajin Simu1_run4.mdcrd 1 100000 10
trajin Simu1_run5.mdcrd 1 100000 10
trajin Simu1_run6.mdcrd 1 100000 10
trajin Simu1_run7.mdcrd 1 100000 10
trajin Simu1_run8.mdcrd 1 100000 10
strip :WAT
strip :Na+
diffusion :1-53,55-67 1.0 average MSDs1tr_without54_WA
go
In this case there are 8 coordinate files of 5ns each that amount to a 40ns MSD simulation. The MSD plot averaged over 4 simulations is presented below.
[cid:bef89445-b394-4ba3-b66a-b72286421824]
Thanks for the help.
Best Regards,
Pau
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
