Date: Tue, 12 Jan 2021 15:52:25 +0900
Dear David and All,
I'm getting the analysis that I wanted, but xmgrace is not working on my
system and the error message looks too much out of my expertise to fix.
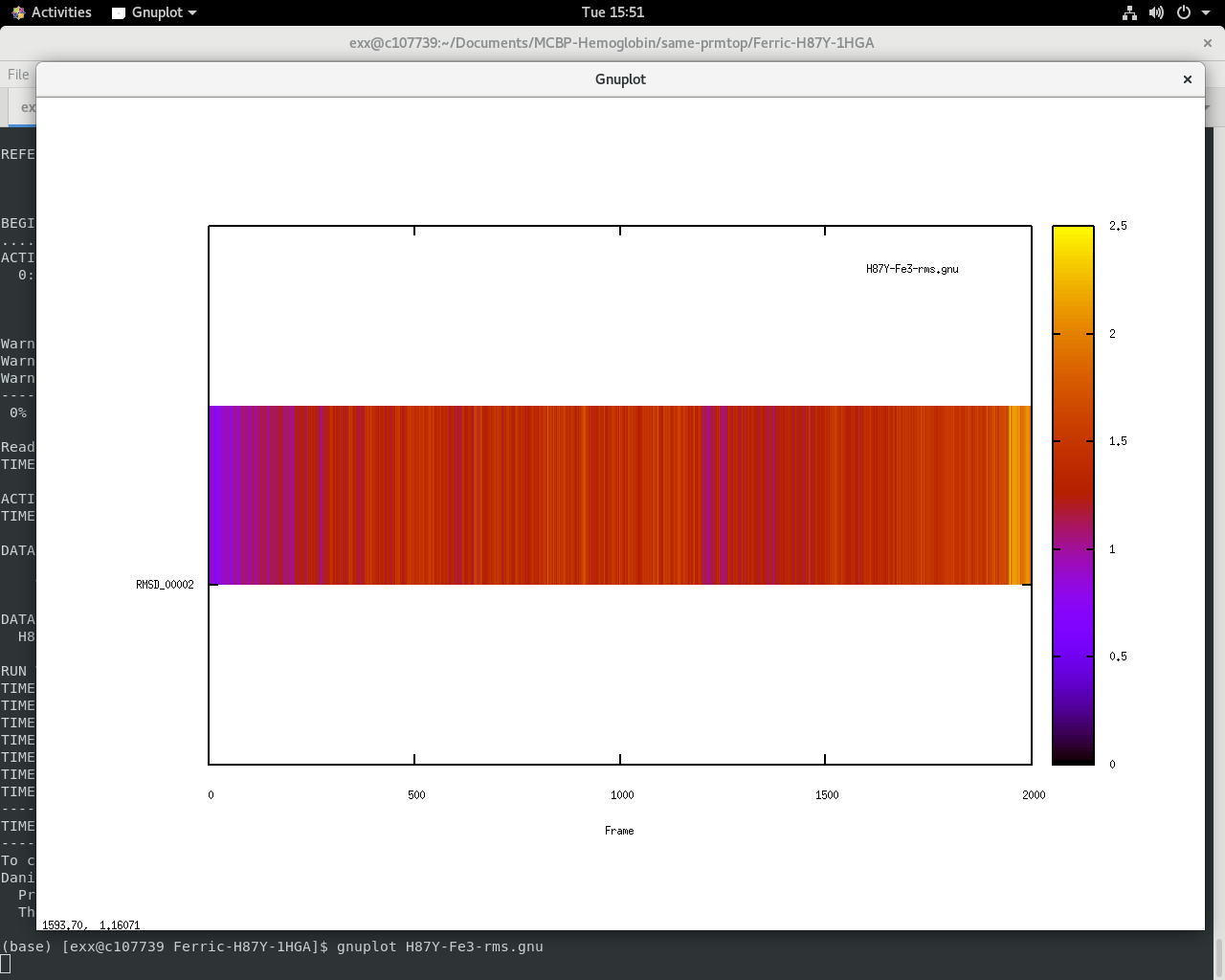
If I choose gnu format for rms out command that I get a display that
doesn't look like a usual rmsd plot (I think it displays 3d plot/map?)
Can you please suggest to me if there is a quick fix to the xmgrace error
or better options to display a line plot in gnuplot?
I understand this is not amber specific but useful suggestions are
highly appreciated.
thank you and best regards
(base) [exx.c107739 test]$ head rms.gnu
set pm3d map corners2color c1
set ytics 1.000, 1.000
set ytics("RMSD_00002" 1.000)
set xlabel "Frame"
set ylabel ""
set yrange [ 0.000: 3.000]
set xrange [ 0.000:2002.000]
splot "-" with pm3d title "H87Y-Fe3-rms.gnu"
1.000 1.000 0.6856
1.000 2.000 0
[image: image.png]
On Mon, Jan 11, 2021 at 10:18 PM David A Case <david.case.rutgers.edu>
wrote:
> On Mon, Jan 11, 2021, Vaibhav Dixit wrote:
>
> >I have simulated two protein structures in which only one residue was
> >mutated and thus backbone sequence is the same.
> >I'm guessing it must be possible to use cpptraj to align the backbone
> >coordinates frame-by-frame, and estimate rmsd to check if the mutants
> >sample the same conformational space.
> >
> >parm prot-top
> >trajin prot.nc
> >reference prot.inpcrd
> >trajin prot1.nc parm
>
> The command you want is "rms", something like this:
>
> rms reference :1-141.C,CA,N out rms.dat
>
> The manual shows lots of keywords that can be added to the rms command, but
> this should get you going.
>
> ....dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
