Date: Mon, 4 Jan 2021 14:26:57 +0530
The cpptraj command doesn't read the correct order of nc files for input,
that command with wildcard read the .nc files as follows:
md1000.nc
md1001.nc
md1002.nc
md1003.nc
md1004.nc
md1005.nc
md1006.nc
md1007.nc
md1008.nc
md1009.nc
md100.nc
md1010.nc
md1011.nc
md1012.nc
md1013.nc
md1014.nc
md1015.nc
md1016.nc
md1017.nc
md1018.nc
md1019.nc
md101.nc and so on...
please suggest the cpptraj command for *combining the 2500 nc files.*
*thanks*
On Thu, Dec 31, 2020 at 3:24 PM Syeda Amna Arshi <seyydaamnaarshi.gmail.com>
wrote:
> Hii,
> first for combining the nc files i used following cpptraj input:
> parm ABC_wation.prmtop
> trajin md*.nc 1 last 10 # load frames in steps of 10
> autoimage # automatic centre
> strip :WAT # strip all residues with name WAT
> trajout ABC_25.nc netcdf ! write out combined trajectory from 20.1ns run
> go
>
>
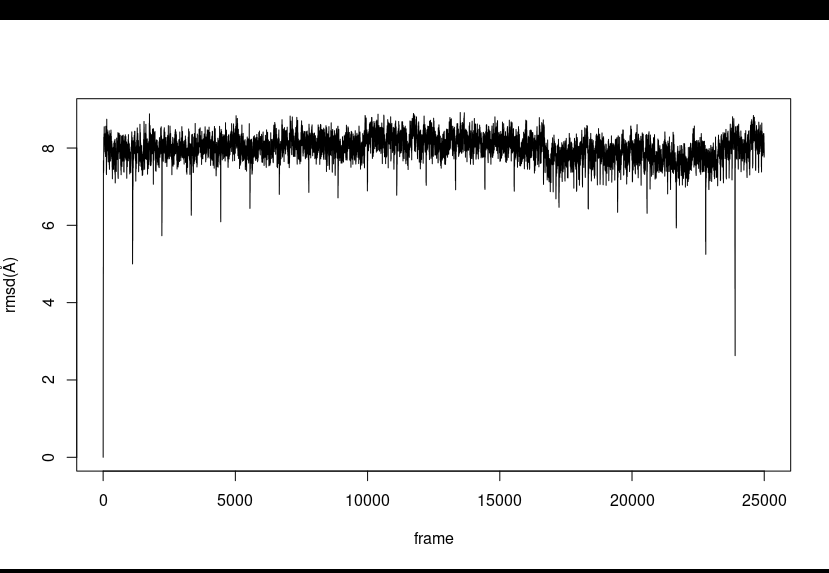
> further for the rmsd:
> parm ABC_ion.prmtop
> trajin ABC_25.nc
> autoimage anchor :1-123
> rms first out rmsd.dat .CA
>
> and the resultant plot is attached below
>
>
> [image: image.png]
>
> On Thu, Dec 24, 2020 at 12:02 PM Syeda Amna Arshi <
> seyydaamnaarshi.gmail.com> wrote:
>
>> I am very confused after autoimage it shows same drops in the rmsd at the
>> same time point. Please look at the input files also may be i have to
>> change the parameters.
>>
>> On Mon, Dec 21, 2020 at 12:39 PM Syeda Amna Arshi <
>> seyydaamnaarshi.gmail.com> wrote:
>>
>>> I think that my complex has been moving out of the octahedron water box
>>> (TIP3PBOX) during the simulation, it looks like part of the complex goes
>>> out of the box at many points. Are there any parameters in my input files
>>> that I should try adjusting?
>>> Here are the settings that I used for my input files
>>>
>>> --complex initial minimization solvent + ions
>>> &cntrl imin = 1,maxcyc = 3000,ncyc = 1500,ntb = 1,ntr =
>>> 1,cut = 10.0/
>>> Hold the Protein + DNA fixed
>>> 100.0
>>> RES 1 153
>>> END
>>> END
>>>
>>> --complex initial minimization solvent + ions
>>> &cntrl imin = 1,maxcyc = 5000,ncyc = 2500,ntb = 1,ntr = 1,cut
>>> = 10.0/
>>> Hold the Protein + DNA fixed
>>> 25.0
>>> RES 1 153
>>> END
>>> END
>>>
>>> --Complex minimization for whole system without restraint
>>> &cntrl imin = 1,maxcyc = 5000,ncyc = 2500,ntb = 1,ntr = 0,cut
>>> = 10.0/
>>>
>>>
>>> --complex: 20ps MD with res on the protein + DNA complex
>>> &cntrl
>>> imin = 0, irest = 0, ntx = 1, ntb = 1, cut = 10.0,
>>> ntr = 1, ntc = 2, ntf = 2, tempi = 0.0, temp0 = 300.0,ntt
>>> = 3, gamma_ln = 1.0, nstlim = 10000, dt = 0.002, ntpr = 10000, ntwx =
>>> 10000, ntwr = 10000,
>>>
>>> /
>>> Keep DNA fixed with weak restraints
>>> 10.0
>>> RES 1 153
>>> END
>>> END
>>>
>>>
>>> --complex: 80ps MD with 0.5 Kcal res on protein + DNA complex
>>> &cntrl
>>> imin = 0, irest = 1, ntx = 7, ntb = 1, cut = 10.0,
>>> ntr = 1, ntc = 2, ntf = 2, tempi = 300.0, temp0 = 300.0,
>>> ntt = 3, gamma_ln = 1.0, nstlim = 40000, dt = 0.002, ntpr = 40000,
>>> ntwx = 40000, ntwr = 40000 ig = -1
>>> /
>>> Keep Protein + DNA fixed with weak restraints
>>> 0.5
>>> RES 1 153
>>> END
>>> END
>>>
>>>
>>>
>>> --complex: 200ps of MD
>>> &cntrl
>>> imin = 0, irest = 1, ntx = 7,
>>> ntb = 2, pres0 = 1.0, ntp = 1,
>>> taup = 2.0,
>>> cut = 10.0, ntr = 0, ig = -1,
>>> ntc = 2, ntf = 2,
>>> tempi = 300.0, temp0 = 300.0,
>>> ntt = 3, gamma_ln = 1.0,
>>> nstlim = 100000, dt = 0.002,
>>> ntpr = 100000, ntwx = 10000, ntwr = 100000
>>> /
>>>
>>>
>>>
>>>
>>>
>>> Sincerely,
>>>
>>> *Syeda Amna Arshi*
>>>
>>> *Ph.D. Scholar*
>>>
>>> Multidisciplinary Centre For Advanced Research and Studies
>>>
>>> Jamia Millia Islamia
>>>
>>> Delhi-110025
>>>
>>> Ph.no: 7737638065
>>>
>>>
>>> [image: Image result for save world by plant][image: Image result for
>>> save world by plant white background]
>>>
>>
>>
>> --
>>
>> Sincerely,
>>
>> *Syeda Amna Arshi*
>>
>> *Ph.D. Scholar*
>>
>> Multidisciplinary Centre For Advanced Research and Studies
>>
>> Jamia Millia Islamia
>>
>> Delhi-110025
>>
>> Ph.no: 7737638065
>>
>>
>> [image: Image result for save world by plant][image: Image result for
>> save world by plant white background]
>>
>
>
> --
>
> Sincerely,
>
> *Syeda Amna Arshi*
>
> *Ph.D. Scholar*
>
> Multidisciplinary Centre For Advanced Research and Studies
>
> Jamia Millia Islamia
>
> Delhi-110025
>
> Ph.no: 7737638065
>
>
> [image: Image result for save world by plant][image: Image result for
> save world by plant white background]
>
-- Sincerely, *Syeda Amna Arshi* *Ph.D. Scholar* Multidisciplinary Centre For Advanced Research and Studies Jamia Millia Islamia Delhi-110025 Ph.no: 7737638065 [image: Image result for save world by plant][image: Image result for save world by plant white background]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
