Date: Fri, 18 Dec 2020 22:17:21 +0530
Dear All,
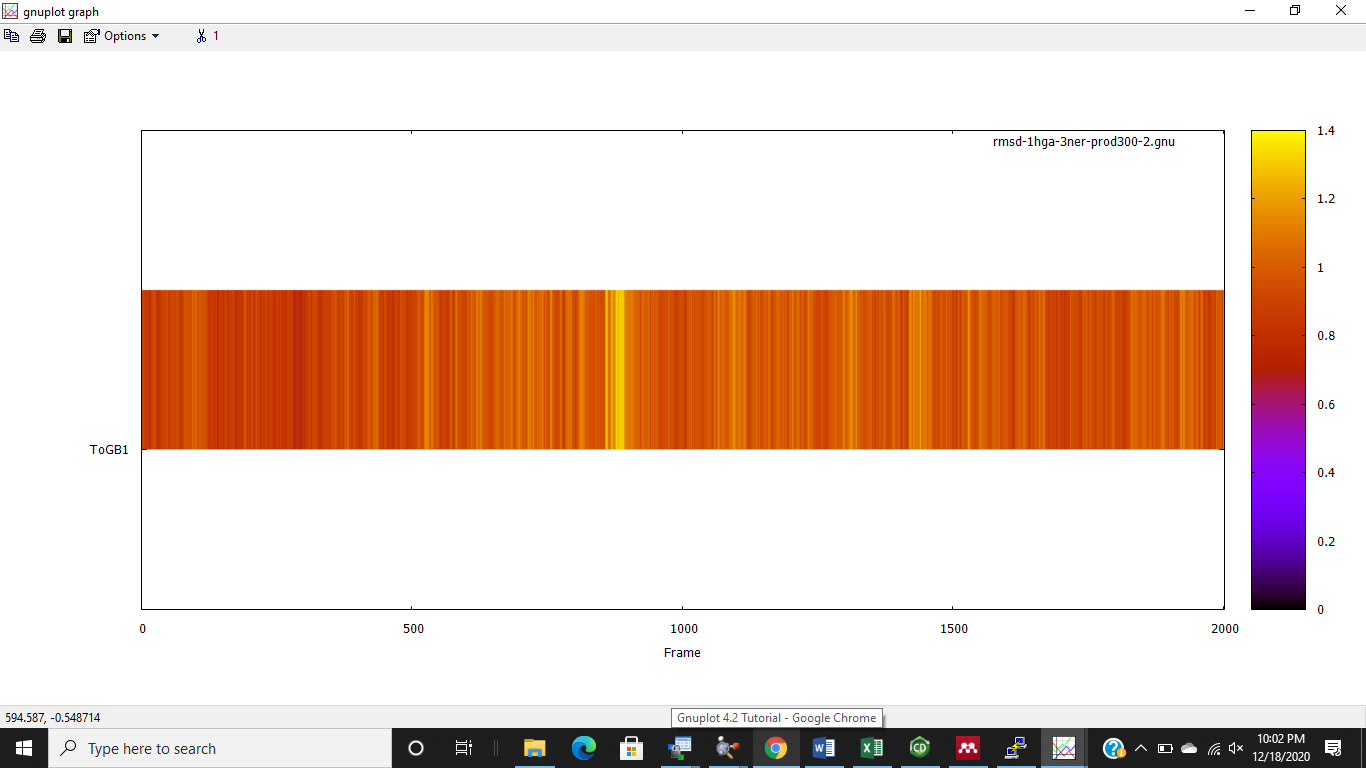
After autoimage, I'm getting meaningful values as shown below (rmsd max
1.4), meaning that the system is well equilibrated and suitable for
analysis, right?
So do you recommend to always use the reimaged trajectory for analysis and
interpretation of conformations, interactions etc (especially when the
default option iwrap=0 was used during MD).
The manual says it has no effect on Es and forces, thus is it safe to
assume that the positive Amber FF Es that I got in other cases was not
related to "not performing" the autoimage before estimating SP Es (and was
related to aspects clarified by other in that thread)?
Looking forward to your valuable suggestions.
thanks and best regards.
[image: image.png]
On Fri, Dec 18, 2020 at 7:46 PM Daniel Roe <daniel.r.roe.gmail.com> wrote:
> Hi,
>
> Carlos is right, what you're seeing (the instantaneous jumps) are
> almost certainly imaging artifacts. The 'autoimage' command in cpptraj
> is usually good at fixing these. E.g.
>
> parm <topology>
> trajin <trajectory>
> autoimage
> rms first <other keywords>
>
> -Dan
>
> On Fri, Dec 18, 2020 at 6:44 AM Vaibhav Dixit <vaibhavadixit.gmail.com>
> wrote:
> >
> > I calculated this in VMD (using RMSD trajectory tool) and used the option
> > to Align.
> > I'm not sure if it is equivalent to reimage in cpptraj.
> > I will try with cpptraj and get back here in case the problem presists.
> > thanks for suggesting.
> >
> > On Fri, Dec 18, 2020 at 4:58 PM Carlos Simmerling <
> > carlos.simmerling.gmail.com> wrote:
> >
> > > I agree that they are likely due to imaging, since they come back down
> > > suddenly too.
> > > did you try to reimage in cpptraj?
> > >
> > > On Fri, Dec 18, 2020 at 6:06 AM Vaibhav Dixit <vaibhavadixit.gmail.com
> >
> > > wrote:
> > >
> > >> Dear All,
> > >> I'm simulating a complex of two metalloproteins (120 ns trajectory).
> > >> After about 80 ns, I'm observing a sudden jump in rmsd which looks
> > >> unphysical but I'm not sure if it is due to some issue with wrapping.
> > >> Visualization in VMD shows that one of the protein suddenly changes
> > >> position.
> > >> Both proteins seem to be well within the solvent box, thus I'm unable
> to
> > >> guess what is the problem here.
> > >> I have pasted two snapshots below in which the sudden jump is
> apparent.
> > >> Please suggest to me ways in which I can check if these jumps are
> genuine
> > >> or some type of artifact in the simulation.
> > >> Thank you and best regards.
> > >>
> > >>
> > >> [image: image.png]
> > >>
> > >> [image: image.png]
> > >>
> > >> [image: image.png]
> > >>
> > >> --
> > >>
> > >> Regards,
> > >>
> > >> Dr. Vaibhav A. Dixit,
> > >>
> > >> Visiting Scientist at the Manchester Institute of Biotechnology
> (MIB), The
> > >> University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
> > >> AND
> > >> Assistant Professor,
> > >> Department of Pharmacy,
> > >> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
> > >> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
> > >> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
> > >> India.
> > >> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
> > >> Email: vaibhav.dixit.pilani.bits-pilani.ac.in,
> vaibhavadixit.gmail.com
> > >> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
> > >> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
> > >>
> > >> ORCID ID: https://orcid.org/0000-0003-4015-2941
> > >>
> > >> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
> > >>
> > >> P Please consider the environment before printing this e-mail
> > >> _______________________________________________
> > >> AMBER mailing list
> > >> AMBER.ambermd.org
> > >> http://lists.ambermd.org/mailman/listinfo/amber
> > >>
> > >
> >
> > --
> >
> > Regards,
> >
> > Dr. Vaibhav A. Dixit,
> >
> > Visiting Scientist at the Manchester Institute of Biotechnology (MIB),
> The
> > University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
> > AND
> > Assistant Professor,
> > Department of Pharmacy,
> > ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
> > Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
> > VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
> > India.
> > Phone No. +91 1596 255652, Mob. No. +91-7709129400,
> > Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
> > http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
> > https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
> >
> > ORCID ID: https://orcid.org/0000-0003-4015-2941
> >
> > http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
> >
> > P Please consider the environment before printing this e-mail
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
