Date: Thu, 14 May 2020 23:27:24 -0500
Hi All,
I am trying to do PCA for the combining of 3 trajectories of a protein with
3 different ligands. Could you please help me to clarify the below points?
1. From the output modes.dat (calculate eigenvalues for all eigenvectors),
my first eigenvector only accounts for 14% of the total motion. It requires
65 eigenvectors to describe 80% of the total motion. Do you think the
results are not good? I suppose that the first PC should contribute more
than 50% and it requires only a few PCs to represent the most dynamic modes
of the concatenating 3 trajectories.
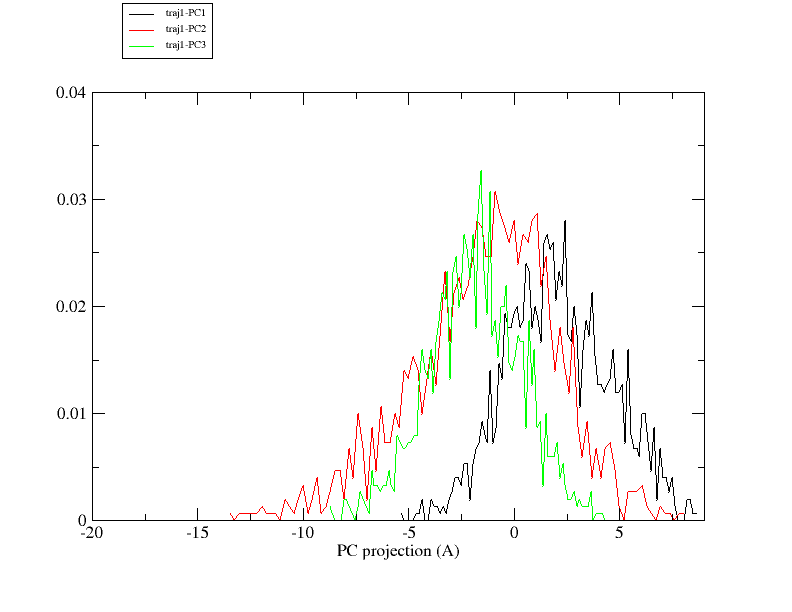
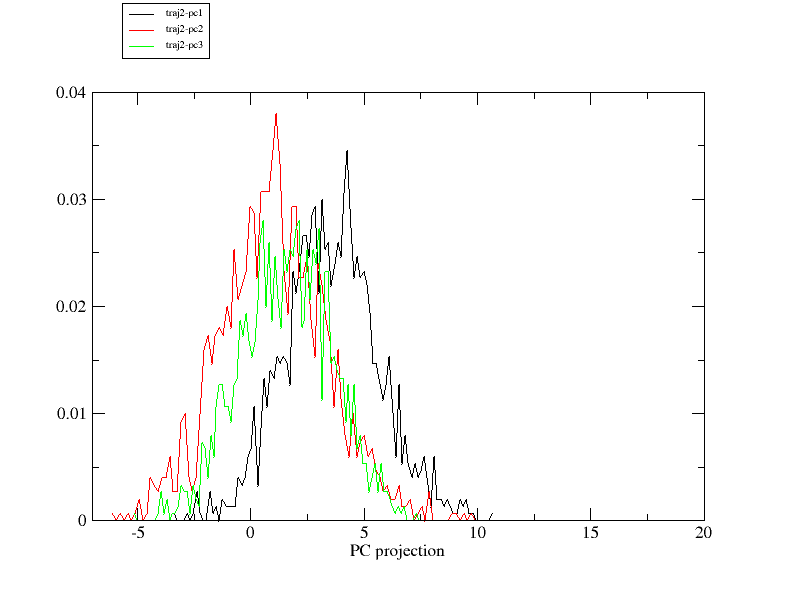
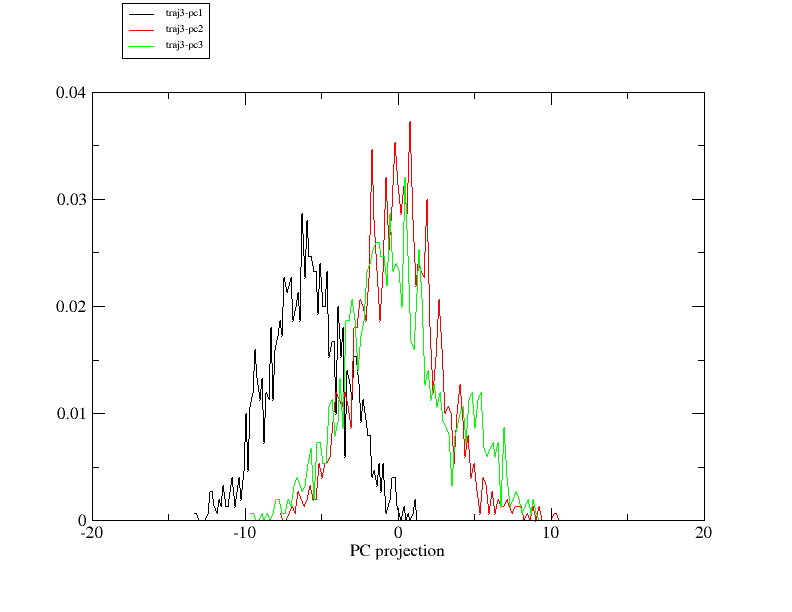
2. I am trying to define pcmin and pcmax to generate the pseudo-trajectory
by checking the histogram of PC projection. I notice that the projection of
the first 3 PCs does not distribute evenly around 0, especially PC1. (png
files are attached). Since I did center my data before diagonalizing the
covariance matrix, I suppose 0 should be the center of all PC projections
from each trajectory. Please correct me if I am wrong.
Below is the script I used based on the PCA tutorial on Amber.
parm topo.prmtop
trajin traj1.nc 500 2000 1
trajin traj2.nc 500 2000 1
trajin traj3.nc 500 2000 1
rms first :1-309.CA
average crdset avg
createcrd avg-traj
run
crdaction avg-traj rms ref avg :1-309.CA
crdaction avg-traj matrix covar :1-309.CA name covar-matrix
runanalysis diagmatrix covar-matrix out evecs.dat vecs 927
crdaction avg-traj projection traj1 modes evecs.dat beg 1 end 927 :1-309.CA
crdframes 1,1501 out traj1-projection.dat
crdaction avg-traj projection traj2 modes evecs.dat beg 1 end 927 :1-309.CA
crdframes 1502,3002 out traj2-projection.dat
crdaction avg-traj projection traj3 modes evecs.dat beg 1 end 927 :1-309.CA
crdframes 3003,last out traj3-projection.dat
hist traj1:1 bins 100 out hist-traj1.agr norm name traj1-pc1
hist traj1:2 bins 100 out hist-traj1.agr norm name traj1-pc2
hist traj1:3 bins 100 out hist-traj1.agr norm name traj1-pc3
hist traj2:1 bins 100 out hist-traj2.agr norm name traj2-pc1
hist traj2:2 bins 100 out hist-traj2.agr norm name traj2-pc2
hist traj2:3 bins 100 out hist-traj2.agr norm name traj2-pc3
hist traj3:1 bins 100 out hist-traj3.agr norm name traj3-pc1
hist traj3:2 bins 100 out hist-traj3.agr norm name traj3-pc2
hist traj3:3 bins 100 out hist-traj3.agr norm name traj3-pc3
run
readdata evecs.dat name Evecs
runanalysis modes name Evecs trajout PC1.pdb pcmin -20 pcmax 20 tmode 1
trajoutmask :1-309.CA trajoutfmt pdb
runanalysis modes name Evecs trajout PC2.pdb pcmin -20 pcmax 20 tmode 2
trajoutmask :1-309.CA trajoutfmt pdb
runanalysis modes name Evecs trajout PC3.pdb pcmin -20 pcmax 20 tmode 3
trajoutmask :1-309.CA trajoutfmt pdb
runanalysis modes name Evecs eigenval out modes.dat
run
3. I would like to filter the motions along PC1/PC2 from the individual
trajectory. Is it the appropriate script to do?
parm topo.prmtop
trajin traj1.nc 500 2000 1
rms first :1-303.CA
average crdset avg
createcrd avg-traj
run
readdata evecs.dat
reaction avg-traj projection myprojection modes evecs.dat out myproject.txt
beg 1 end 3 :1-303.CA
filter myprojection:1 min 0 max 4 out filter.dat
trajout filter-traj1-pc1.pdb
Again, since the histogram of PC1 projection from traj1 is not center at 0,
can I set my min and max as 0 and 4, respectively?
Thank you for your help
Kat
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: traj1-proj.png)
(image/png attachment: traj2-proj.png)
(image/png attachment: traj3-proj.png)
