Date: Fri, 17 Apr 2020 00:58:54 -0500
Hi Lachele,
Thank you for your reply.
I have build the hemicellulose using glycam carbohydrate builder and then I
am trying to load the pdb file of ''hemicellulose'' using loadpdb command
in xleap.
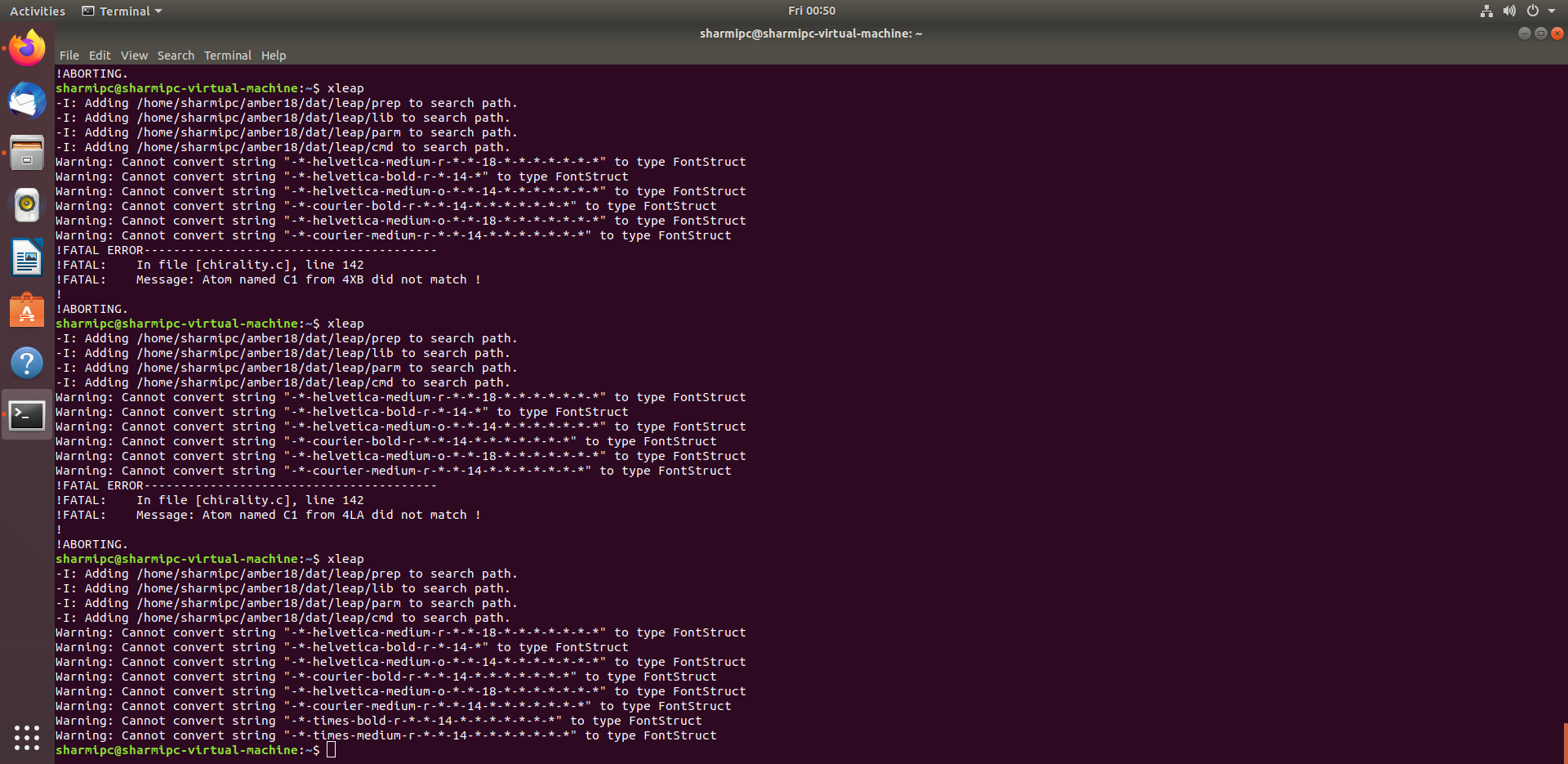
But, It shows the error !FATAL: Message: Atom named C1 from 4LA did not
match !
Attached is the screenshot.
Do you have any idea why this is happening?
Any help would be appreciated.
Thank you again.
Sincerely,
Pinky
On Thu, Apr 16, 2020 at 3:23 AM Lachele Foley <lf.list.gmail.com> wrote:
> I usually use tleap rather than xleap, but you should be able to use
> either. See my previous reply, but also consider the 'combine'
> command.
>
> Learning to use xleap or tleap takes a little time, but the Amber
> manual does a pretty good job of describing the functions.
>
> On Wed, Apr 15, 2020 at 11:14 PM Pinky Mazumder <pmazumder67.gmail.com>
> wrote:
> >
> > Dear Amber Users,
> >
> > I have built cellulose chain using xleap command. On the other hand,
> > I have made the chain of hemicellulose using GLYCAM_06 carbohydrate
> builder.
> >
> > Now, I need to put the chain close together.
> >
> >
> > So, my question is, how can I make the both chains closer together?
> Should
> > I load the chain of hemicellulose in xleap? If it is, then what is the
> > command for this job?
> >
> >
> > Any help would be appreciated. Please forgive me if I am asking something
> > silly.
> >
> > Thank you.
> >
> > --
> > Pinky
> > AL,US
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
>
>
>
> --
> :-) Lachele
> Lachele Foley
> CCRC/UGA
> Athens, GA USA
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
-- Pinky AL,US
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_from_2020-04-17_00-50-31.png)
