Date: Mon, 13 Jan 2020 14:26:41 +0000
Get Outlook for Android<https://aka.ms/ghei36>
________________________________
From: Tiejun Wei <t.wei.qmul.ac.uk>
Sent: Monday, January 13, 2020 1:30:24 PM
To: amber.ambermd.org <amber.ambermd.org>
Subject: Fw: Periodic box problem when simulating Membrane system
Tiejun Wei
<http://www.qmul.ac.uk/lifesciences/research/phd/index.html>CSC<http://www.sbcs.qmul.ac.uk/postgraduate/research/studentships/china-scholarship-council/> PhD student
Computational Biology/Biophysics
<https://bessantlab.org/>DuffyLab<http://www.duffylab.co.uk/> (QMUL)
[http://www.daviddang.uk/icon/linkedin-sig.png]<https://www.linkedin.com/in/tiejunwei/>
________________________________
From: Tiejun Wei
Sent: Monday, January 13, 2020 1:28 PM
To: amber.ambermd.org <amber.ambermd.org>
Subject: Periodic box problem when simulating Membrane system
Hi AMBER users:
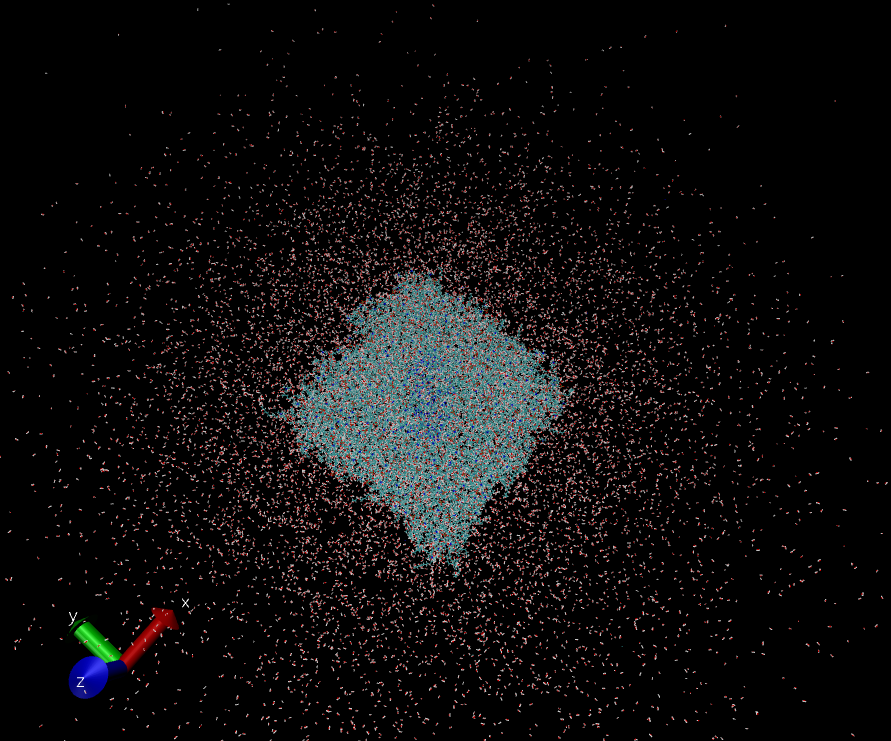
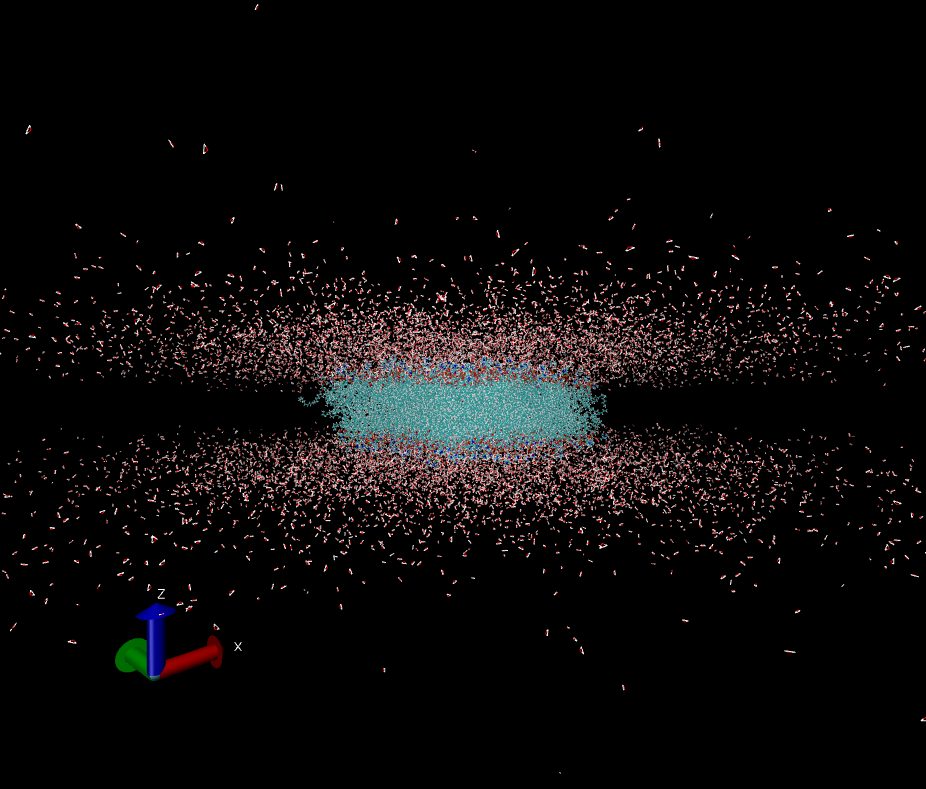
I'm using AMBER to simulate a membrane system and found my TIP3P water molecules are "spreading". (see pics attached)
The system was generated by CHARMM-GUI but I'm not using auto-generated constraint files.
I'm not particularly interested in protein structural change, we focus more on the co-factors of the protein. Just curious, is this normal?
If not, what's the correct scenario looks like? is there any examples of AMBER/NAMD config files for membrane simulation?
Best,
Tiejun
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_from_2020-01-13_13-23-32.png)
(image/png attachment: Screenshot_from_2020-01-13_13-23-26.png)
