Date: Sun, 5 Jan 2020 19:48:43 +0530
Hiii
Attached are my files.two perl scripts for two different runs. And fasta
file for sequence..
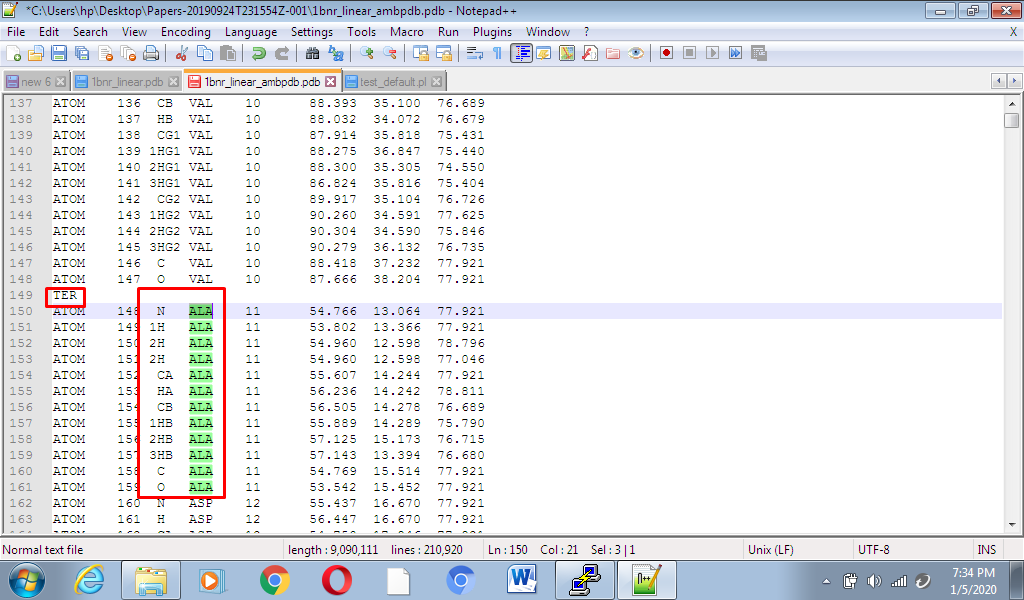
Exactly pdb file generated through tleap had TER before ALA.
Moreover ALA had hydrogens in it.. See the attached screen shots and files.
Pls suggest how to deal with this..
On Sun 5 Jan, 2020, 7:18 PM David Case, <david.case.rutgers.edu> wrote:
> On Sun, Jan 05, 2020, ankita mehta wrote:
>
> >Why tleap is considering all ALANINE to be NALA?
>
> We have so little information about what you did, and what pdb file you
> loaded, to offer any useful information. You apparently used a sequence
> command, but we don't have any information about what step that was, or
> what was in it.
>
> Generally, the default in tleap is to make change an N-terminal ALA
> residue to NALA. Note that NALA (and other N-terminal residues) does
> not have an "H" atom in it. Do you have any unwanted TER cards in you
> PDB file right before the ALA residues?
>
> ....dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
- text/plain attachment: 1bnr.fasta.txt
(image/png attachment: Screenshot_8.png)
- text/x-perl attachment: test_default.pl
- text/x-perl attachment: test_box.pl
