Date: Thu, 26 Sep 2019 10:13:53 +0800
Thanks for sharing the information and publications!
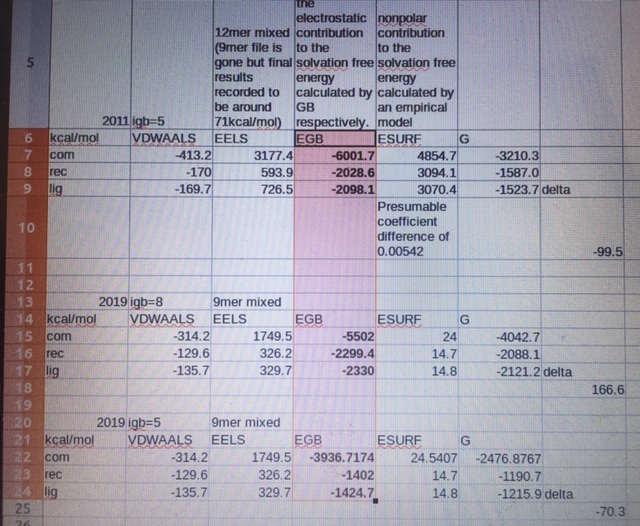
I see that the absolute solvation energies for a 12mer RNA duplex is between -6000 to -6800 kcal/mol, and my 9mer duplex is near -5500kcal/mol, pretty consistent with each other. The base sequences are not the same. Running PBSAs will now be on my list.
I used mbond3 for the prmtop file and trajectory generation, and now I made another mbond2 prmtop file, that gave no difference from the mbond3, either with igb=5 or 8 during the MMGBSA energy calculation.
Out of analysis interest I put the results in a spreadsheet file and I’ve taken a photo:
(Resized to smaller photo or it can’t automatically send through)
The direct cause of this difference seems to be that the GB solvation energy on the single strands (rec and lig, both ssRNA) are proportionally more favorable in the igb=8 run vs the igb=5 run, in turn making the formation of the complex less favorable (bold and highlighted cells).
>> On Sep 26, 2019, at 12:13 AM, Hai Nguyen <nhai.qn.gmail.com> wrote:
>>
>> On Wed, Sep 25, 2019 at 11:18 AM Liao <liaojunzhuo.aliyun.com> wrote:
>>
>> Thanks there Professor Case,
>
>
>> As I found out that igb=5 and igb=8 made a huge difference, though I’m yet
>> to find out why.
>
> which radii set (mbondi2, bondi, ...) you used for your prmtop file?
> igb=8 used mbondi2 for parameterization.
>
> Hai
>
>
>> Yes doing it by hand is essential, though I still need to figure out
>> sander’s commands’ compatibility between old and new Amber-some of them
>> seem to be incompatible, at least from my preliminary try. Still checking
>> Amber 19’s manual for the newest by-hand MMGBSA examples.
>>
>>>> On Sep 25, 2019, at 9:54 PM, David Case <david.case.rutgers.edu> wrote:
>>>>
>>>> On Wed, Sep 25, 2019, Liao wrote:
>>>>
>>>> The issue is that I’ve ran the exact same molecule before with AMBER11,
>>>> and the binding energy result was very reasonable, at -71 kcal/mol. The
>>>> only significant system setup difference I can think of is that back
>> with
>>>> AMBER 11 there was no charge neutralization via Tleap with Na+ for the
>>>> solvated complex, and now with AMBER 19 22 Na+ were added to neutralize
>>>> the system.
>>>
>>> The way to proceed here is to run MM-PBSA "by hand" so that you know
>>> exactly how things like ions are being handled. (See the coments at the
>>> beginning of Chap. 32 in the Reference Manual.)
>>>
>>> The "classic" implicit solvent approach is to consider mobile ions like
>>> Na+ as the part of the solvent, that is, to strip then out before doing
>>> any analysis. It's not clear (to me) how well alternative approaches
>>> that try to treat ions as a part of the RNA may be expected to work.
>>> Nor do I know (although others on this list may know) how various
>>> automated scripts between Amber11 and Amber19 might have handled this.
>>> Hence the recommendation to make sure you understand in detail exactly
>>> what the automated scripts are going.
>>>
>>>>
>>>> From the current energy results, the G for the gas phase dsRNA complex
>>>> seems to be quite high (compared to the 2 single strands) like as if the
>>>> negative charges on RNA are repulsing the 2 strands away.
>>>
>>> This is exactly what I would expect to be happening. Solvent screening
>>> (from both dielectric effects and from mobile counterions) will
>>> mostly cancel this electrostatic repulsion.
>>>
>>> ....good luck....dac
>>>
>>> _______________________________________________
>>> AMBER mailing list
>>> AMBER.ambermd.org
>>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: image1.jpeg)
