Date: Mon, 29 Apr 2019 13:05:27 +0200
Hi Amber folks,
I would like to ask you people for help.
Namely, I’ve run an umbrella COM PMF calculation on DMPC72-H4OBBZ system
(ligand attached). Everything went smoothly. I run 32 windows for 30 ns long
traj. The only thing was the negative windows which I’ve turned to positive
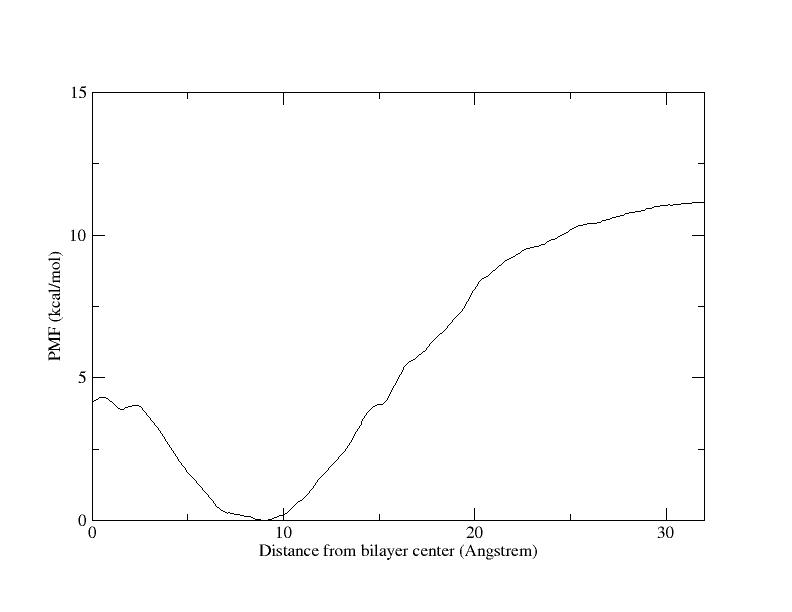
with Callum’s “fix..” script. However, when I calculated wham free
energies the plot looks unusual (see data and pic). If I’ve learnt well the
PMF line tends to be around zero while we approach to 32Å (is that right?).
As You can see I have maximum plateau at 32Å equal to ca. 11 kcal/mol. In
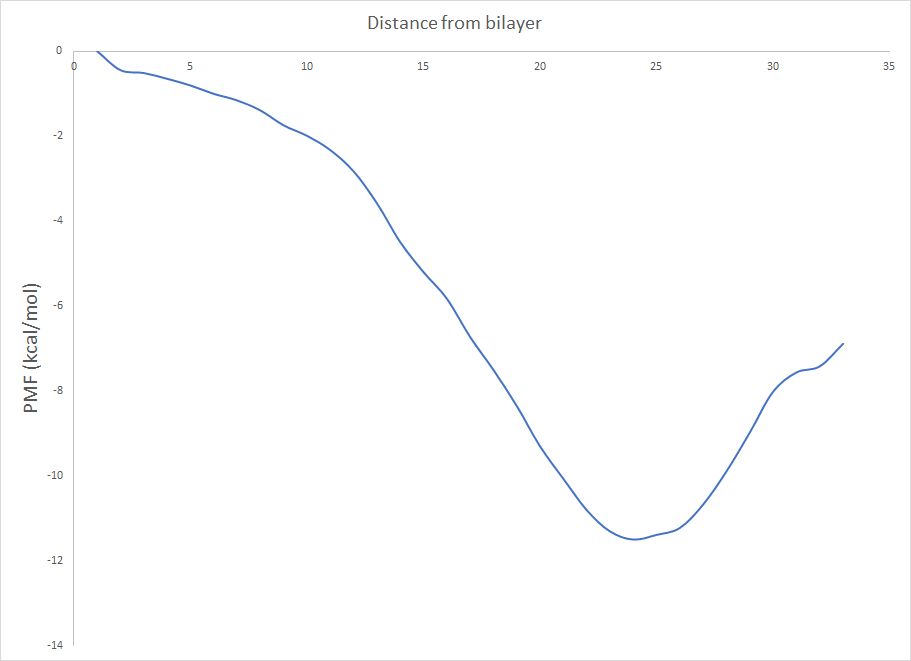
addition, the second part of pmf.out contains two columns named as “#Window
Free”
While my past PMF calculation showed similarity between these two plots now
its quite different.
Would You be so kind and help me as how to understand this.
Best wishes,
Zoran
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: h4obbz_ac.jpg)
- application/octet-stream attachment: plot_free_energy.dat
- application/octet-stream attachment: plot_free_energy.agr
(image/jpeg attachment: plot_free_energy.jpg)
(image/jpeg attachment: plot_free_energy_windows_free.jpg)
