Date: Wed, 18 Jul 2018 12:21:08 +0530
Dear Amber/GLYCAM users,
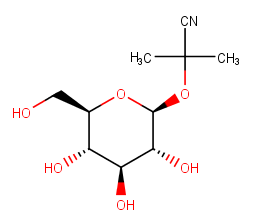
I am trying to simulate below mentioned ligand (linamarin) docked to
b-Glucoside enzyme. It is made up of a glucose moiety in which anomeric O-H
is substituted by a cyanogenic moitey. I would like to know if one would
necessarily require the use GLYCAM force-field to model such system. If so
how to prepare the PDB/mol2 file to comply with GLYCAM naming conventions.
Regards,
-- D L Senal Dinuka Grad.Chem., A.I.Chem.C. Research Assistant College of Chemical Sciences Institute of Chemistry Ceylon Rajagiriya Sri Lanka +94 77 627 4678
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
