Date: Thu, 12 Jul 2018 22:44:36 +0000 (UTC)
Dear All,
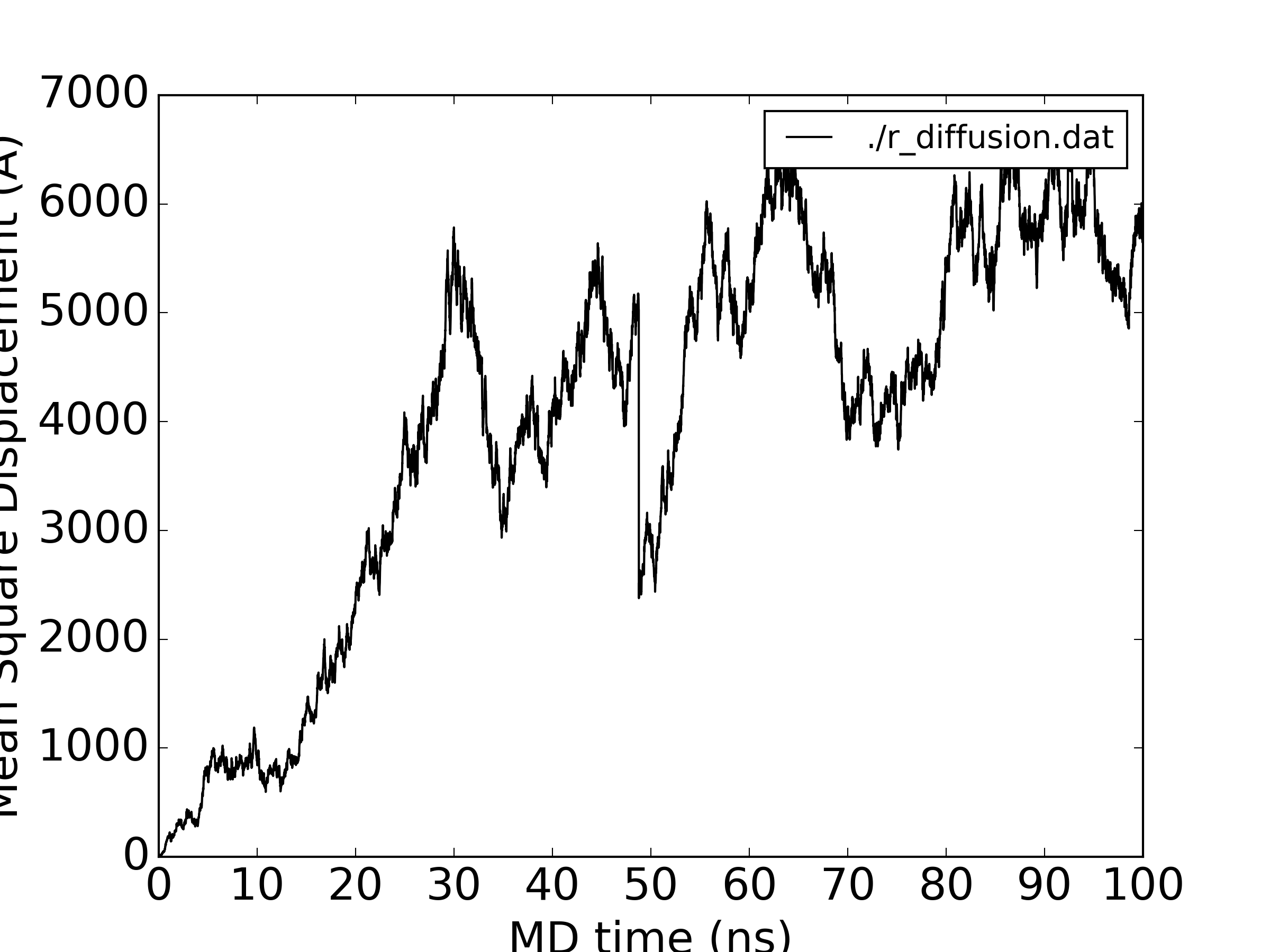
I have bring up a topic that is widely discussed on the list in the past, yet I do not seem to get it right. I would like derive translational diffusion coefficient from Mean Squared Displacements. The simulations were run with iwrap =1 (see protocol below), I processed the trajectories in cpptraj with unwrap and tried to get the diffusion coefficient (second script). I was expecting that the plots the MSDs (overall, total, x,y,z) should be monotonously increasing, but this is not the case (see attached). At around 50ns there is a clear downward spike in all plots, which does not make sense. Accordingly the fitted diffusion coefficients are sort of random (certainly do not match the experimental data) and in some cases are even negative. Not using the noimage keyword does produce slightly different MSD plots with the same issues. Could anyone help me figuring out where it goes wrong?
Thanks for any idea!Kris
Production
&cntrl
imin=0, ntx=5, irest=1,
ntb=2,cut=8.0,
ntp=1, pres0=1.0,
ntc=2,
ntf=2,
ntt=3, temp0=310.0, gamma_ln=1.0,
nstlim=5000000, dt=0.002,
ioutfm=1,
iwrap=1, ioutfm=1,
ntpr=5000, ntwr=50000, ntwx=5000,
ntr=0, ig=-1,
/
- cpptraj script for trajectory processing
parm ../../dna_w.prmtop
trajin ../../5_prodNPT/dna_prod1.netcdf
trajin ../../5_prodNPT/dna_prod2.netcdf
trajin ../../5_prodNPT/dna_prod3.netcdf
trajin ../../5_prodNPT/dna_prod4.netcdf
trajin ../../5_prodNPT/dna_prod5.netcdf
trajin ../../5_prodNPT/dna_prod6.netcdf
trajin ../../5_prodNPT/dna_prod7.netcdf
trajin ../../5_prodNPT/dna_prod8.netcdf
trajin ../../5_prodNPT/dna_prod9.netcdf
trajin ../../5_prodNPT/dna_prod10.netcdf
unwrap
diffusion :1-20 separateout diffusion.dat noimage diffout DiffusionConstants.lst
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: r_diffusion.png)
